Liang Zhan
for the Alzheimer's Disease Neuroimaging Initiative
Deep Models, Shallow Alignment: Uncovering the Granularity Mismatch in Neural Decoding
Jan 29, 2026Abstract:Neural visual decoding is a central problem in brain computer interface research, aiming to reconstruct human visual perception and to elucidate the structure of neural representations. However, existing approaches overlook a fundamental granularity mismatch between human and machine vision, where deep vision models emphasize semantic invariance by suppressing local texture information, whereas neural signals preserve an intricate mixture of low-level visual attributes and high-level semantic content. To address this mismatch, we propose Shallow Alignment, a novel contrastive learning strategy that aligns neural signals with intermediate representations of visual encoders rather than their final outputs, thereby striking a better balance between low-level texture details and high-level semantic features. Extensive experiments across multiple benchmarks demonstrate that Shallow Alignment significantly outperforms standard final-layer alignment, with performance gains ranging from 22% to 58% across diverse vision backbones. Notably, our approach effectively unlocks the scaling law in neural visual decoding, enabling decoding performance to scale predictably with the capacity of pre-trained vision backbones. We further conduct systematic empirical analyses to shed light on the mechanisms underlying the observed performance gains.
Aligning Findings with Diagnosis: A Self-Consistent Reinforcement Learning Framework for Trustworthy Radiology Reporting
Jan 06, 2026Abstract:Multimodal Large Language Models (MLLMs) have shown strong potential for radiology report generation, yet their clinical translation is hindered by architectural heterogeneity and the prevalence of factual hallucinations. Standard supervised fine-tuning often fails to strictly align linguistic outputs with visual evidence, while existing reinforcement learning approaches struggle with either prohibitive computational costs or limited exploration. To address these challenges, we propose a comprehensive framework for self-consistent radiology report generation. First, we conduct a systematic evaluation to identify optimal vision encoder and LLM backbone configurations for medical imaging. Building on this foundation, we introduce a novel "Reason-then-Summarize" architecture optimized via Group Relative Policy Optimization (GRPO). This framework restructures generation into two distinct components: a think block for detailed findings and an answer block for structured disease labels. By utilizing a multi-dimensional composite reward function, we explicitly penalize logical discrepancies between the generated narrative and the final diagnosis. Extensive experiments on the MIMIC-CXR benchmark demonstrate that our method achieves state-of-the-art performance in clinical efficacy metrics and significantly reduces hallucinations compared to strong supervised baselines.
R-GenIMA: Integrating Neuroimaging and Genetics with Interpretable Multimodal AI for Alzheimer's Disease Progression
Dec 22, 2025Abstract:Early detection of Alzheimer's disease (AD) requires models capable of integrating macro-scale neuroanatomical alterations with micro-scale genetic susceptibility, yet existing multimodal approaches struggle to align these heterogeneous signals. We introduce R-GenIMA, an interpretable multimodal large language model that couples a novel ROI-wise vision transformer with genetic prompting to jointly model structural MRI and single nucleotide polymorphisms (SNPs) variations. By representing each anatomically parcellated brain region as a visual token and encoding SNP profiles as structured text, the framework enables cross-modal attention that links regional atrophy patterns to underlying genetic factors. Applied to the ADNI cohort, R-GenIMA achieves state-of-the-art performance in four-way classification across normal cognition (NC), subjective memory concerns (SMC), mild cognitive impairment (MCI), and AD. Beyond predictive accuracy, the model yields biologically meaningful explanations by identifying stage-specific brain regions and gene signatures, as well as coherent ROI-Gene association patterns across the disease continuum. Attention-based attribution revealed genes consistently enriched for established GWAS-supported AD risk loci, including APOE, BIN1, CLU, and RBFOX1. Stage-resolved neuroanatomical signatures identified shared vulnerability hubs across disease stages alongside stage-specific patterns: striatal involvement in subjective decline, frontotemporal engagement during prodromal impairment, and consolidated multimodal network disruption in AD. These results demonstrate that interpretable multimodal AI can synthesize imaging and genetics to reveal mechanistic insights, providing a foundation for clinically deployable tools that enable earlier risk stratification and inform precision therapeutic strategies in Alzheimer's disease.
Why Text Prevails: Vision May Undermine Multimodal Medical Decision Making
Dec 15, 2025Abstract:With the rapid progress of large language models (LLMs), advanced multimodal large language models (MLLMs) have demonstrated impressive zero-shot capabilities on vision-language tasks. In the biomedical domain, however, even state-of-the-art MLLMs struggle with basic Medical Decision Making (MDM) tasks. We investigate this limitation using two challenging datasets: (1) three-stage Alzheimer's disease (AD) classification (normal, mild cognitive impairment, dementia), where category differences are visually subtle, and (2) MIMIC-CXR chest radiograph classification with 14 non-mutually exclusive conditions. Our empirical study shows that text-only reasoning consistently outperforms vision-only or vision-text settings, with multimodal inputs often performing worse than text alone. To mitigate this, we explore three strategies: (1) in-context learning with reason-annotated exemplars, (2) vision captioning followed by text-only inference, and (3) few-shot fine-tuning of the vision tower with classification supervision. These findings reveal that current MLLMs lack grounded visual understanding and point to promising directions for improving multimodal decision making in healthcare.
Conditional Neural ODE for Longitudinal Parkinson's Disease Progression Forecasting
Nov 06, 2025Abstract:Parkinson's disease (PD) shows heterogeneous, evolving brain-morphometry patterns. Modeling these longitudinal trajectories enables mechanistic insight, treatment development, and individualized 'digital-twin' forecasting. However, existing methods usually adopt recurrent neural networks and transformer architectures, which rely on discrete, regularly sampled data while struggling to handle irregular and sparse magnetic resonance imaging (MRI) in PD cohorts. Moreover, these methods have difficulty capturing individual heterogeneity including variations in disease onset, progression rate, and symptom severity, which is a hallmark of PD. To address these challenges, we propose CNODE (Conditional Neural ODE), a novel framework for continuous, individualized PD progression forecasting. The core of CNODE is to model morphological brain changes as continuous temporal processes using a neural ODE model. In addition, we jointly learn patient-specific initial time and progress speed to align individual trajectories into a shared progression trajectory. We validate CNODE on the Parkinson's Progression Markers Initiative (PPMI) dataset. Experimental results show that our method outperforms state-of-the-art baselines in forecasting longitudinal PD progression.
DRE: An Effective Dual-Refined Method for Integrating Small and Large Language Models in Open-Domain Dialogue Evaluation
Jun 04, 2025Abstract:Large Language Models (LLMs) excel at many tasks but struggle with ambiguous scenarios where multiple valid responses exist, often yielding unreliable results. Conversely, Small Language Models (SLMs) demonstrate robustness in such scenarios but are susceptible to misleading or adversarial inputs. We observed that LLMs handle negative examples effectively, while SLMs excel with positive examples. To leverage their complementary strengths, we introduce SLIDE (Small and Large Integrated for Dialogue Evaluation), a method integrating SLMs and LLMs via adaptive weighting. Building on SLIDE, we further propose a Dual-Refinement Evaluation (DRE) method to enhance SLM-LLM integration: (1) SLM-generated insights guide the LLM to produce initial evaluations; (2) SLM-derived adjustments refine the LLM's scores for improved accuracy. Experiments demonstrate that DRE outperforms existing methods, showing stronger alignment with human judgment across diverse benchmarks. This work illustrates how combining small and large models can yield more reliable evaluation tools, particularly for open-ended tasks such as dialogue evaluation.
Zeus: Zero-shot LLM Instruction for Union Segmentation in Multimodal Medical Imaging
Apr 09, 2025Abstract:Medical image segmentation has achieved remarkable success through the continuous advancement of UNet-based and Transformer-based foundation backbones. However, clinical diagnosis in the real world often requires integrating domain knowledge, especially textual information. Conducting multimodal learning involves visual and text modalities shown as a solution, but collecting paired vision-language datasets is expensive and time-consuming, posing significant challenges. Inspired by the superior ability in numerous cross-modal tasks for Large Language Models (LLMs), we proposed a novel Vision-LLM union framework to address the issues. Specifically, we introduce frozen LLMs for zero-shot instruction generation based on corresponding medical images, imitating the radiology scanning and report generation process. {To better approximate real-world diagnostic processes}, we generate more precise text instruction from multimodal radiology images (e.g., T1-w or T2-w MRI and CT). Based on the impressive ability of semantic understanding and rich knowledge of LLMs. This process emphasizes extracting special features from different modalities and reunion the information for the ultimate clinical diagnostic. With generated text instruction, our proposed union segmentation framework can handle multimodal segmentation without prior collected vision-language datasets. To evaluate our proposed method, we conduct comprehensive experiments with influential baselines, the statistical results and the visualized case study demonstrate the superiority of our novel method.}
End-to-End Deep Learning for Structural Brain Imaging: A Unified Framework
Feb 23, 2025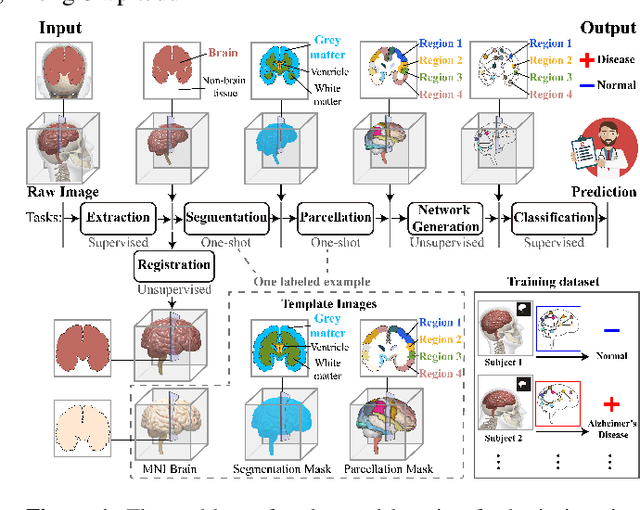
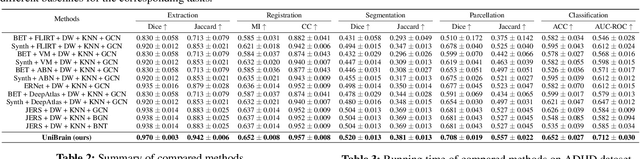
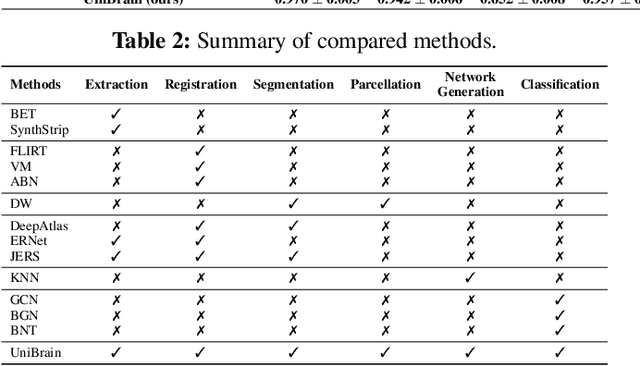
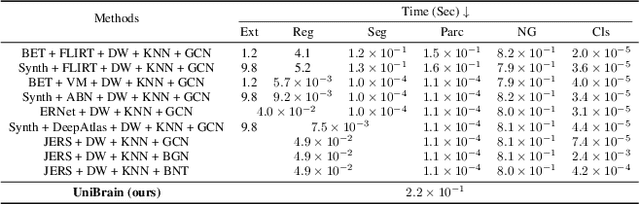
Abstract:Brain imaging analysis is fundamental in neuroscience, providing valuable insights into brain structure and function. Traditional workflows follow a sequential pipeline-brain extraction, registration, segmentation, parcellation, network generation, and classification-treating each step as an independent task. These methods rely heavily on task-specific training data and expert intervention to correct intermediate errors, making them particularly burdensome for high-dimensional neuroimaging data, where annotations and quality control are costly and time-consuming. We introduce UniBrain, a unified end-to-end framework that integrates all processing steps into a single optimization process, allowing tasks to interact and refine each other. Unlike traditional approaches that require extensive task-specific annotations, UniBrain operates with minimal supervision, leveraging only low-cost labels (i.e., classification and extraction) and a single labeled atlas. By jointly optimizing extraction, registration, segmentation, parcellation, network generation, and classification, UniBrain enhances both accuracy and computational efficiency while significantly reducing annotation effort. Experimental results demonstrate its superiority over existing methods across multiple tasks, offering a more scalable and reliable solution for neuroimaging analysis. Our code and data can be found at https://github.com/Anonymous7852/UniBrain
Rethinking Functional Brain Connectome Analysis: Do Graph Deep Learning Models Help?
Jan 28, 2025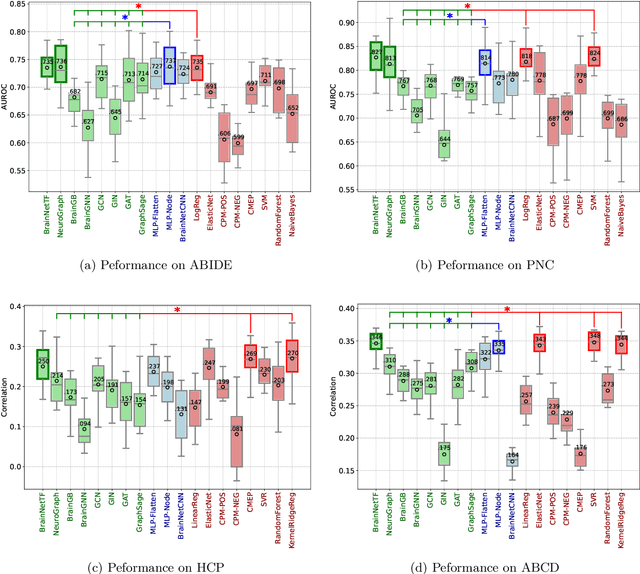
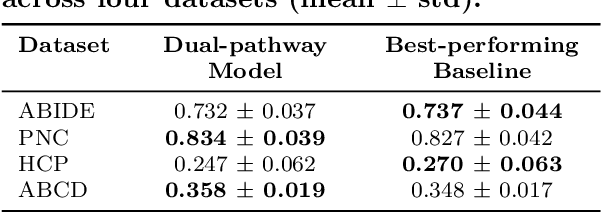
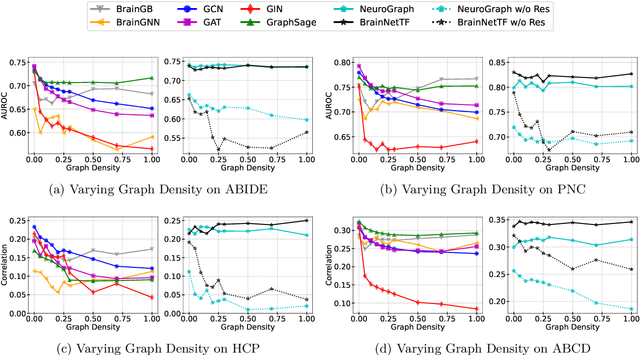
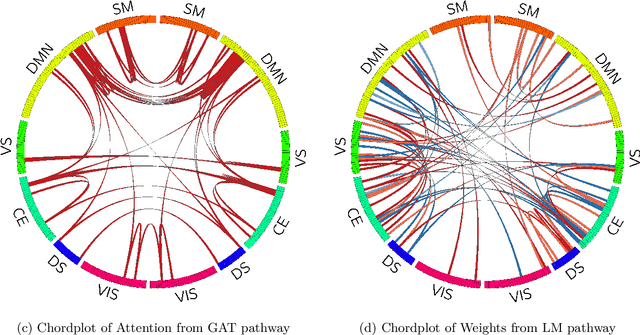
Abstract:Functional brain connectome is crucial for deciphering the neural mechanisms underlying cognitive functions and neurological disorders. Graph deep learning models have recently gained tremendous popularity in this field. However, their actual effectiveness in modeling the brain connectome remains unclear. In this study, we re-examine graph deep learning models based on four large-scale neuroimaging studies encompassing diverse cognitive and clinical outcomes. Surprisingly, we find that the message aggregation mechanism, a hallmark of graph deep learning models, does not help with predictive performance as typically assumed, but rather consistently degrades it. To address this issue, we propose a hybrid model combining a linear model with a graph attention network through dual pathways, achieving robust predictions and enhanced interpretability by revealing both localized and global neural connectivity patterns. Our findings urge caution in adopting complex deep learning models for functional brain connectome analysis, emphasizing the need for rigorous experimental designs to establish tangible performance gains and perhaps more importantly, to pursue improvements in model interpretability.
A Self-guided Multimodal Approach to Enhancing Graph Representation Learning for Alzheimer's Diseases
Dec 09, 2024



Abstract:Graph neural networks (GNNs) are powerful machine learning models designed to handle irregularly structured data. However, their generic design often proves inadequate for analyzing brain connectomes in Alzheimer's Disease (AD), highlighting the need to incorporate domain knowledge for optimal performance. Infusing AD-related knowledge into GNNs is a complicated task. Existing methods typically rely on collaboration between computer scientists and domain experts, which can be both time-intensive and resource-demanding. To address these limitations, this paper presents a novel self-guided, knowledge-infused multimodal GNN that autonomously incorporates domain knowledge into the model development process. Our approach conceptualizes domain knowledge as natural language and introduces a specialized multimodal GNN capable of leveraging this uncurated knowledge to guide the learning process of the GNN, such that it can improve the model performance and strengthen the interpretability of the predictions. To evaluate our framework, we curated a comprehensive dataset of recent peer-reviewed papers on AD and integrated it with multiple real-world AD datasets. Experimental results demonstrate the ability of our method to extract relevant domain knowledge, provide graph-based explanations for AD diagnosis, and improve the overall performance of the GNN. This approach provides a more scalable and efficient alternative to inject domain knowledge for AD compared with the manual design from the domain expert, advancing both prediction accuracy and interpretability in AD diagnosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge