Runxue Bao
Safe Screening Rules for Group SLOPE
Jun 11, 2025Abstract:Variable selection is a challenging problem in high-dimensional sparse learning, especially when group structures exist. Group SLOPE performs well for the adaptive selection of groups of predictors. However, the block non-separable group effects in Group SLOPE make existing methods either invalid or inefficient. Consequently, Group SLOPE tends to incur significant computational costs and memory usage in practical high-dimensional scenarios. To overcome this issue, we introduce a safe screening rule tailored for the Group SLOPE model, which efficiently identifies inactive groups with zero coefficients by addressing the block non-separable group effects. By excluding these inactive groups during training, we achieve considerable gains in computational efficiency and memory usage. Importantly, the proposed screening rule can be seamlessly integrated into existing solvers for both batch and stochastic algorithms. Theoretically, we establish that our screening rule can be safely employed with existing optimization algorithms, ensuring the same results as the original approaches. Experimental results confirm that our method effectively detects inactive feature groups and significantly boosts computational efficiency without compromising accuracy.
Any Large Language Model Can Be a Reliable Judge: Debiasing with a Reasoning-based Bias Detector
May 21, 2025



Abstract:LLM-as-a-Judge has emerged as a promising tool for automatically evaluating generated outputs, but its reliability is often undermined by potential biases in judgment. Existing efforts to mitigate these biases face key limitations: in-context learning-based methods fail to address rooted biases due to the evaluator's limited capacity for self-reflection, whereas fine-tuning is not applicable to all evaluator types, especially closed-source models. To address this challenge, we introduce the Reasoning-based Bias Detector (RBD), which is a plug-in module that identifies biased evaluations and generates structured reasoning to guide evaluator self-correction. Rather than modifying the evaluator itself, RBD operates externally and engages in an iterative process of bias detection and feedback-driven revision. To support its development, we design a complete pipeline consisting of biased dataset construction, supervision collection, distilled reasoning-based fine-tuning of RBD, and integration with LLM evaluators. We fine-tune four sizes of RBD models, ranging from 1.5B to 14B, and observe consistent performance improvements across all scales. Experimental results on 4 bias types--verbosity, position, bandwagon, and sentiment--evaluated using 8 LLM evaluators demonstrate RBD's strong effectiveness. For example, the RBD-8B model improves evaluation accuracy by an average of 18.5% and consistency by 10.9%, and surpasses prompting-based baselines and fine-tuned judges by 12.8% and 17.2%, respectively. These results highlight RBD's effectiveness and scalability. Additional experiments further demonstrate its strong generalization across biases and domains, as well as its efficiency.
Safe Screening Rules for Group OWL Models
Apr 08, 2025Abstract:Group Ordered Weighted $L_{1}$-Norm (Group OWL) regularized models have emerged as a useful procedure for high-dimensional sparse multi-task learning with correlated features. Proximal gradient methods are used as standard approaches to solving Group OWL models. However, Group OWL models usually suffer huge computational costs and memory usage when the feature size is large in the high-dimensional scenario. To address this challenge, in this paper, we are the first to propose the safe screening rule for Group OWL models by effectively tackling the structured non-separable penalty, which can quickly identify the inactive features that have zero coefficients across all the tasks. Thus, by removing the inactive features during the training process, we may achieve substantial computational gain and memory savings. More importantly, the proposed screening rule can be directly integrated with the existing solvers both in the batch and stochastic settings. Theoretically, we prove our screening rule is safe and also can be safely applied to the existing iterative optimization algorithms. Our experimental results demonstrate that our screening rule can effectively identify the inactive features and leads to a significant computational speedup without any loss of accuracy.
A Self-guided Multimodal Approach to Enhancing Graph Representation Learning for Alzheimer's Diseases
Dec 09, 2024



Abstract:Graph neural networks (GNNs) are powerful machine learning models designed to handle irregularly structured data. However, their generic design often proves inadequate for analyzing brain connectomes in Alzheimer's Disease (AD), highlighting the need to incorporate domain knowledge for optimal performance. Infusing AD-related knowledge into GNNs is a complicated task. Existing methods typically rely on collaboration between computer scientists and domain experts, which can be both time-intensive and resource-demanding. To address these limitations, this paper presents a novel self-guided, knowledge-infused multimodal GNN that autonomously incorporates domain knowledge into the model development process. Our approach conceptualizes domain knowledge as natural language and introduces a specialized multimodal GNN capable of leveraging this uncurated knowledge to guide the learning process of the GNN, such that it can improve the model performance and strengthen the interpretability of the predictions. To evaluate our framework, we curated a comprehensive dataset of recent peer-reviewed papers on AD and integrated it with multiple real-world AD datasets. Experimental results demonstrate the ability of our method to extract relevant domain knowledge, provide graph-based explanations for AD diagnosis, and improve the overall performance of the GNN. This approach provides a more scalable and efficient alternative to inject domain knowledge for AD compared with the manual design from the domain expert, advancing both prediction accuracy and interpretability in AD diagnosis.
Dynamic Uncertainty Ranking: Enhancing In-Context Learning for Long-Tail Knowledge in LLMs
Oct 31, 2024Abstract:Large language models (LLMs) can learn vast amounts of knowledge from diverse domains during pre-training. However, long-tail knowledge from specialized domains is often scarce and underrepresented, rarely appearing in the models' memorization. Prior work has shown that in-context learning (ICL) with retriever augmentation can help LLMs better capture long-tail knowledge, reducing their reliance on pre-trained data. Despite these advances, we observe that LLM predictions for long-tail questions remain uncertain to variations in retrieved samples. To take advantage of the uncertainty in ICL for guiding LLM predictions toward correct answers on long-tail samples, we propose a reinforcement learning-based dynamic uncertainty ranking method for ICL that accounts for the varying impact of each retrieved sample on LLM predictions. Our approach prioritizes more informative and stable samples while demoting misleading ones, updating rankings based on the feedback from the LLM w.r.t. each retrieved sample. To enhance training efficiency and reduce query costs, we introduce a learnable dynamic ranking threshold, adjusted when the model encounters negative prediction shifts. Experimental results on various question-answering datasets from different domains show that our method outperforms the best baseline by $2.76\%$, with a notable $5.96\%$ boost in accuracy on long-tail questions that elude zero-shot inference.
Transfer Learning with Clinical Concept Embeddings from Large Language Models
Sep 20, 2024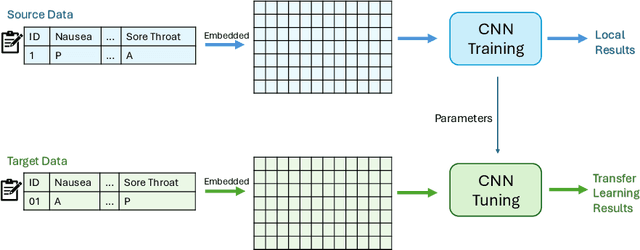

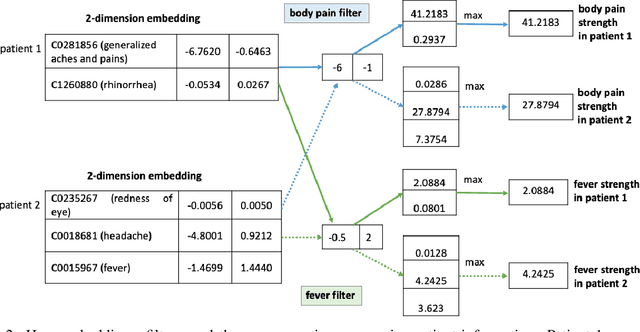

Abstract:Knowledge sharing is crucial in healthcare, especially when leveraging data from multiple clinical sites to address data scarcity, reduce costs, and enable timely interventions. Transfer learning can facilitate cross-site knowledge transfer, but a major challenge is heterogeneity in clinical concepts across different sites. Large Language Models (LLMs) show significant potential of capturing the semantic meaning of clinical concepts and reducing heterogeneity. This study analyzed electronic health records from two large healthcare systems to assess the impact of semantic embeddings from LLMs on local, shared, and transfer learning models. Results indicate that domain-specific LLMs, such as Med-BERT, consistently outperform in local and direct transfer scenarios, while generic models like OpenAI embeddings require fine-tuning for optimal performance. However, excessive tuning of models with biomedical embeddings may reduce effectiveness, emphasizing the need for balance. This study highlights the importance of domain-specific embeddings and careful model tuning for effective knowledge transfer in healthcare.
Unlocking Memorization in Large Language Models with Dynamic Soft Prompting
Sep 20, 2024



Abstract:Pretrained large language models (LLMs) have revolutionized natural language processing (NLP) tasks such as summarization, question answering, and translation. However, LLMs pose significant security risks due to their tendency to memorize training data, leading to potential privacy breaches and copyright infringement. Accurate measurement of this memorization is essential to evaluate and mitigate these potential risks. However, previous attempts to characterize memorization are constrained by either using prefixes only or by prepending a constant soft prompt to the prefixes, which cannot react to changes in input. To address this challenge, we propose a novel method for estimating LLM memorization using dynamic, prefix-dependent soft prompts. Our approach involves training a transformer-based generator to produce soft prompts that adapt to changes in input, thereby enabling more accurate extraction of memorized data. Our method not only addresses the limitations of previous methods but also demonstrates superior performance in diverse experimental settings compared to state-of-the-art techniques. In particular, our method can achieve the maximum relative improvement of 112.75% and 32.26% over the vanilla baseline in terms of discoverable memorization rate for the text generation task and code generation task respectively.
Pruning as a Domain-specific LLM Extractor
May 10, 2024



Abstract:Large Language Models (LLMs) have exhibited remarkable proficiency across a wide array of NLP tasks. However, the escalation in model size also engenders substantial deployment costs. While few efforts have explored model pruning techniques to reduce the size of LLMs, they mainly center on general or task-specific weights. This leads to suboptimal performance due to lacking specificity on the target domain or generality on different tasks when applied to domain-specific challenges. This work introduces an innovative unstructured dual-pruning methodology, D-Pruner, for domain-specific compression on LLM. It extracts a compressed, domain-specific, and task-agnostic LLM by identifying LLM weights that are pivotal for general capabilities, like linguistic capability and multi-task solving, and domain-specific knowledge. More specifically, we first assess general weight importance by quantifying the error incurred upon their removal with the help of an open-domain calibration dataset. Then, we utilize this general weight importance to refine the training loss, so that it preserves generality when fitting into a specific domain. Moreover, by efficiently approximating weight importance with the refined training loss on a domain-specific calibration dataset, we obtain a pruned model emphasizing generality and specificity. Our comprehensive experiments across various tasks in healthcare and legal domains show the effectiveness of D-Pruner in domain-specific compression. Our code is available at https://github.com/psunlpgroup/D-Pruner.
Auto-Train-Once: Controller Network Guided Automatic Network Pruning from Scratch
Mar 21, 2024



Abstract:Current techniques for deep neural network (DNN) pruning often involve intricate multi-step processes that require domain-specific expertise, making their widespread adoption challenging. To address the limitation, the Only-Train-Once (OTO) and OTOv2 are proposed to eliminate the need for additional fine-tuning steps by directly training and compressing a general DNN from scratch. Nevertheless, the static design of optimizers (in OTO) can lead to convergence issues of local optima. In this paper, we proposed the Auto-Train-Once (ATO), an innovative network pruning algorithm designed to automatically reduce the computational and storage costs of DNNs. During the model training phase, our approach not only trains the target model but also leverages a controller network as an architecture generator to guide the learning of target model weights. Furthermore, we developed a novel stochastic gradient algorithm that enhances the coordination between model training and controller network training, thereby improving pruning performance. We provide a comprehensive convergence analysis as well as extensive experiments, and the results show that our approach achieves state-of-the-art performance across various model architectures (including ResNet18, ResNet34, ResNet50, ResNet56, and MobileNetv2) on standard benchmark datasets (CIFAR-10, CIFAR-100, and ImageNet).
InfuserKI: Enhancing Large Language Models with Knowledge Graphs via Infuser-Guided Knowledge Integration
Feb 18, 2024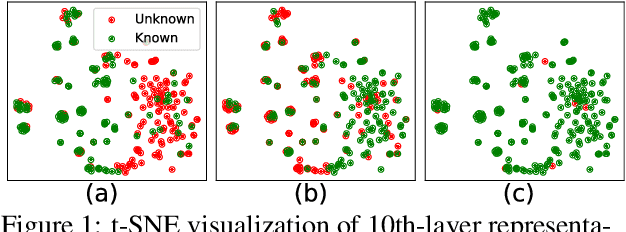
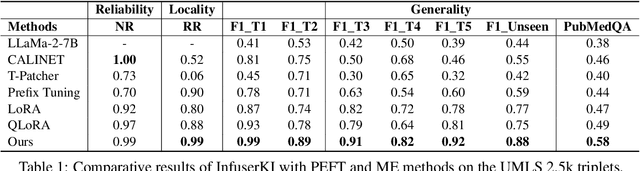
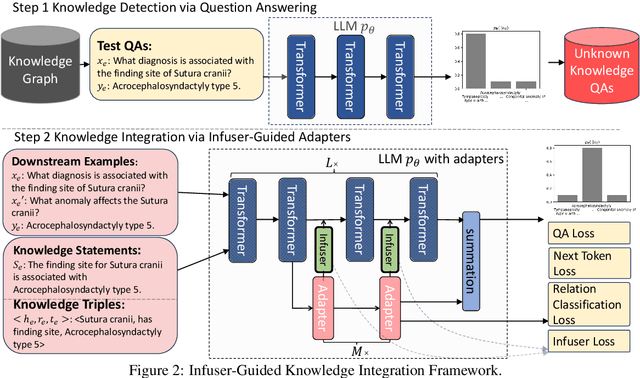
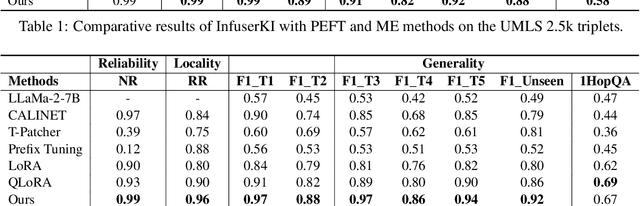
Abstract:Though Large Language Models (LLMs) have shown remarkable open-generation capabilities across diverse domains, they struggle with knowledge-intensive tasks. To alleviate this issue, knowledge integration methods have been proposed to enhance LLMs with domain-specific knowledge graphs using external modules. However, they suffer from data inefficiency as they require both known and unknown knowledge for fine-tuning. Thus, we study a novel problem of integrating unknown knowledge into LLMs efficiently without unnecessary overlap of known knowledge. Injecting new knowledge poses the risk of forgetting previously acquired knowledge. To tackle this, we propose a novel Infuser-Guided Knowledge Integration (InfuserKI) framework that utilizes transformer internal states to determine whether to enhance the original LLM output with additional information, thereby effectively mitigating knowledge forgetting. Evaluations on the UMLS-2.5k and MetaQA domain knowledge graphs demonstrate that InfuserKI can effectively acquire new knowledge and outperform state-of-the-art baselines by 9% and 6%, respectively, in reducing knowledge forgetting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge