Taha Kass-Hout
Focus on What Matters: Enhancing Medical Vision-Language Models with Automatic Attention Alignment Tuning
May 24, 2025Abstract:Medical Large Vision-Language Models (Med-LVLMs) often exhibit suboptimal attention distribution on visual inputs, leading to hallucinated or inaccurate outputs. Existing mitigation methods primarily rely on inference-time interventions, which are limited in attention adaptation or require additional supervision. To address this, we propose A$^3$Tune, a novel fine-tuning framework for Automatic Attention Alignment Tuning. A$^3$Tune leverages zero-shot weak labels from SAM, refines them into prompt-aware labels using BioMedCLIP, and then selectively modifies visually-critical attention heads to improve alignment while minimizing interference. Additionally, we introduce a A$^3$MoE module, enabling adaptive parameter selection for attention tuning across diverse prompts and images. Extensive experiments on medical VQA and report generation benchmarks show that A$^3$Tune outperforms state-of-the-art baselines, achieving enhanced attention distributions and performance in Med-LVLMs.
Any Large Language Model Can Be a Reliable Judge: Debiasing with a Reasoning-based Bias Detector
May 21, 2025



Abstract:LLM-as-a-Judge has emerged as a promising tool for automatically evaluating generated outputs, but its reliability is often undermined by potential biases in judgment. Existing efforts to mitigate these biases face key limitations: in-context learning-based methods fail to address rooted biases due to the evaluator's limited capacity for self-reflection, whereas fine-tuning is not applicable to all evaluator types, especially closed-source models. To address this challenge, we introduce the Reasoning-based Bias Detector (RBD), which is a plug-in module that identifies biased evaluations and generates structured reasoning to guide evaluator self-correction. Rather than modifying the evaluator itself, RBD operates externally and engages in an iterative process of bias detection and feedback-driven revision. To support its development, we design a complete pipeline consisting of biased dataset construction, supervision collection, distilled reasoning-based fine-tuning of RBD, and integration with LLM evaluators. We fine-tune four sizes of RBD models, ranging from 1.5B to 14B, and observe consistent performance improvements across all scales. Experimental results on 4 bias types--verbosity, position, bandwagon, and sentiment--evaluated using 8 LLM evaluators demonstrate RBD's strong effectiveness. For example, the RBD-8B model improves evaluation accuracy by an average of 18.5% and consistency by 10.9%, and surpasses prompting-based baselines and fine-tuned judges by 12.8% and 17.2%, respectively. These results highlight RBD's effectiveness and scalability. Additional experiments further demonstrate its strong generalization across biases and domains, as well as its efficiency.
Enhancing SAM with Efficient Prompting and Preference Optimization for Semi-supervised Medical Image Segmentation
Mar 06, 2025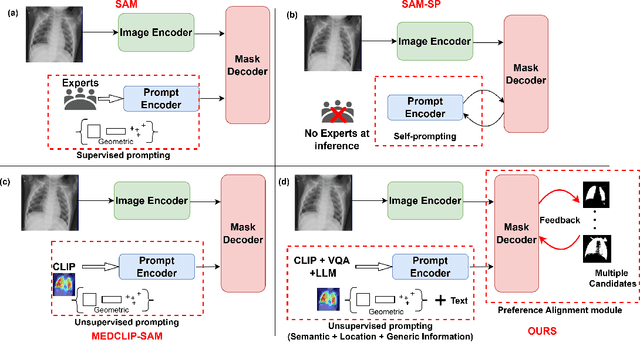
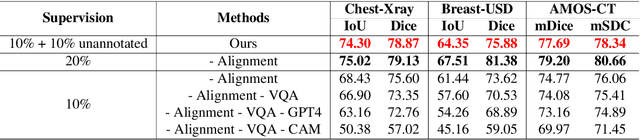
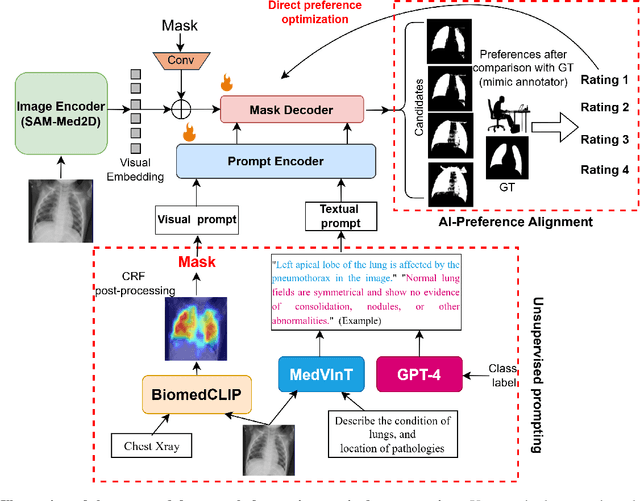
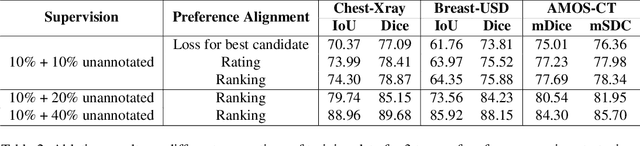
Abstract:Foundational models such as the Segment Anything Model (SAM) are gaining traction in medical imaging segmentation, supporting multiple downstream tasks. However, such models are supervised in nature, still relying on large annotated datasets or prompts supplied by experts. Conventional techniques such as active learning to alleviate such limitations are limited in scope and still necessitate continuous human involvement and complex domain knowledge for label refinement or establishing reward ground truth. To address these challenges, we propose an enhanced Segment Anything Model (SAM) framework that utilizes annotation-efficient prompts generated in a fully unsupervised fashion, while still capturing essential semantic, location, and shape information through contrastive language-image pretraining and visual question answering. We adopt the direct preference optimization technique to design an optimal policy that enables the model to generate high-fidelity segmentations with simple ratings or rankings provided by a virtual annotator simulating the human annotation process. State-of-the-art performance of our framework in tasks such as lung segmentation, breast tumor segmentation, and organ segmentation across various modalities, including X-ray, ultrasound, and abdominal CT, justifies its effectiveness in low-annotation data scenarios.
MedHEval: Benchmarking Hallucinations and Mitigation Strategies in Medical Large Vision-Language Models
Mar 04, 2025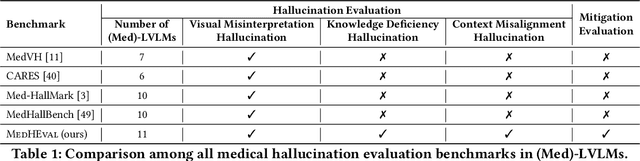
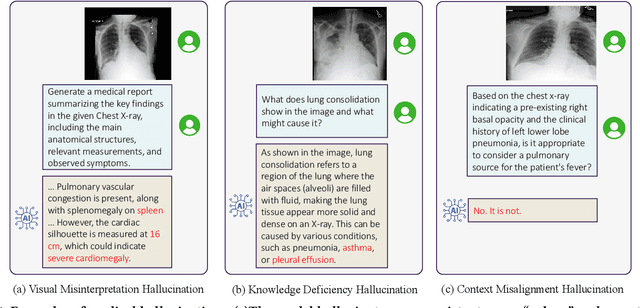
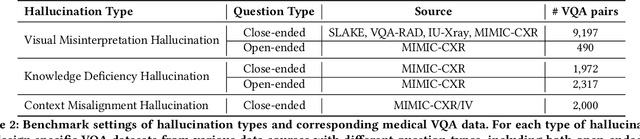
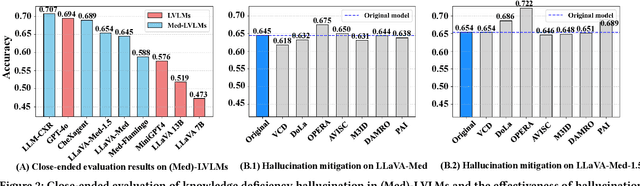
Abstract:Large Vision Language Models (LVLMs) are becoming increasingly important in the medical domain, yet Medical LVLMs (Med-LVLMs) frequently generate hallucinations due to limited expertise and the complexity of medical applications. Existing benchmarks fail to effectively evaluate hallucinations based on their underlying causes and lack assessments of mitigation strategies. To address this gap, we introduce MedHEval, a novel benchmark that systematically evaluates hallucinations and mitigation strategies in Med-LVLMs by categorizing them into three underlying causes: visual misinterpretation, knowledge deficiency, and context misalignment. We construct a diverse set of close- and open-ended medical VQA datasets with comprehensive evaluation metrics to assess these hallucination types. We conduct extensive experiments across 11 popular (Med)-LVLMs and evaluate 7 state-of-the-art hallucination mitigation techniques. Results reveal that Med-LVLMs struggle with hallucinations arising from different causes while existing mitigation methods show limited effectiveness, especially for knowledge- and context-based errors. These findings underscore the need for improved alignment training and specialized mitigation strategies to enhance Med-LVLMs' reliability. MedHEval establishes a standardized framework for evaluating and mitigating medical hallucinations, guiding the development of more trustworthy Med-LVLMs.
Deep Linear Hawkes Processes
Dec 27, 2024Abstract:Marked temporal point processes (MTPPs) are used to model sequences of different types of events with irregular arrival times, with broad applications ranging from healthcare and social networks to finance. We address shortcomings in existing point process models by drawing connections between modern deep state-space models (SSMs) and linear Hawkes processes (LHPs), culminating in an MTPP that we call the deep linear Hawkes process (DLHP). The DLHP modifies the linear differential equations in deep SSMs to be stochastic jump differential equations, akin to LHPs. After discretizing, the resulting recurrence can be implemented efficiently using a parallel scan. This brings parallelism and linear scaling to MTPP models. This contrasts with attention-based MTPPs, which scale quadratically, and RNN-based MTPPs, which do not parallelize across the sequence length. We show empirically that DLHPs match or outperform existing models across a broad range of metrics on eight real-world datasets. Our proposed DLHP model is the first instance of the unique architectural capabilities of SSMs being leveraged to construct a new class of MTPP models.
Dynamic Uncertainty Ranking: Enhancing In-Context Learning for Long-Tail Knowledge in LLMs
Oct 31, 2024Abstract:Large language models (LLMs) can learn vast amounts of knowledge from diverse domains during pre-training. However, long-tail knowledge from specialized domains is often scarce and underrepresented, rarely appearing in the models' memorization. Prior work has shown that in-context learning (ICL) with retriever augmentation can help LLMs better capture long-tail knowledge, reducing their reliance on pre-trained data. Despite these advances, we observe that LLM predictions for long-tail questions remain uncertain to variations in retrieved samples. To take advantage of the uncertainty in ICL for guiding LLM predictions toward correct answers on long-tail samples, we propose a reinforcement learning-based dynamic uncertainty ranking method for ICL that accounts for the varying impact of each retrieved sample on LLM predictions. Our approach prioritizes more informative and stable samples while demoting misleading ones, updating rankings based on the feedback from the LLM w.r.t. each retrieved sample. To enhance training efficiency and reduce query costs, we introduce a learnable dynamic ranking threshold, adjusted when the model encounters negative prediction shifts. Experimental results on various question-answering datasets from different domains show that our method outperforms the best baseline by $2.76\%$, with a notable $5.96\%$ boost in accuracy on long-tail questions that elude zero-shot inference.
Reasoning-Enhanced Healthcare Predictions with Knowledge Graph Community Retrieval
Oct 06, 2024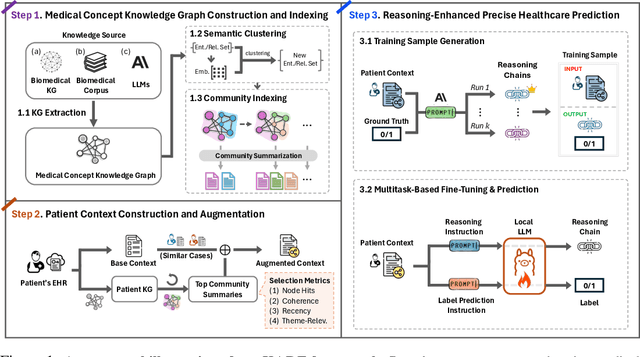
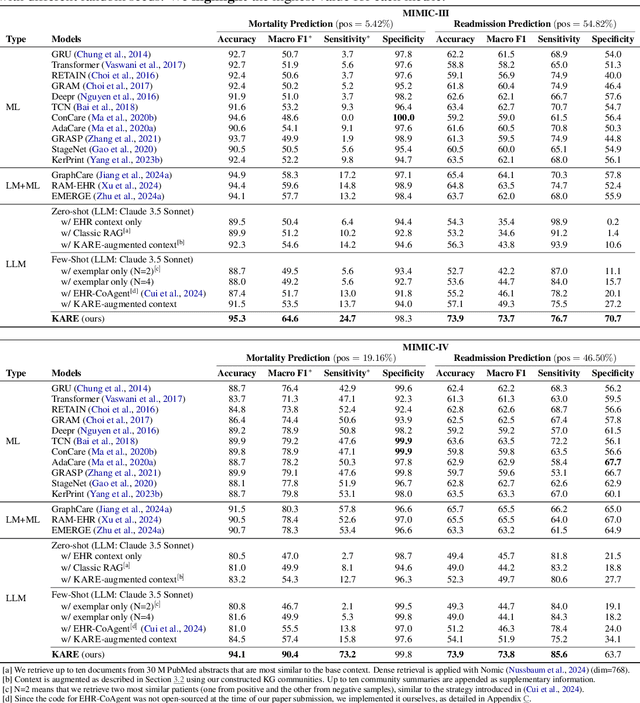
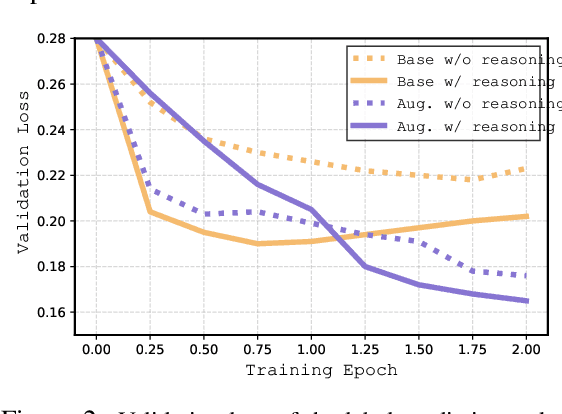
Abstract:Large language models (LLMs) have demonstrated significant potential in clinical decision support. Yet LLMs still suffer from hallucinations and lack fine-grained contextual medical knowledge, limiting their high-stake healthcare applications such as clinical diagnosis. Traditional retrieval-augmented generation (RAG) methods attempt to address these limitations but frequently retrieve sparse or irrelevant information, undermining prediction accuracy. We introduce KARE, a novel framework that integrates knowledge graph (KG) community-level retrieval with LLM reasoning to enhance healthcare predictions. KARE constructs a comprehensive multi-source KG by integrating biomedical databases, clinical literature, and LLM-generated insights, and organizes it using hierarchical graph community detection and summarization for precise and contextually relevant information retrieval. Our key innovations include: (1) a dense medical knowledge structuring approach enabling accurate retrieval of relevant information; (2) a dynamic knowledge retrieval mechanism that enriches patient contexts with focused, multi-faceted medical insights; and (3) a reasoning-enhanced prediction framework that leverages these enriched contexts to produce both accurate and interpretable clinical predictions. Extensive experiments demonstrate that KARE outperforms leading models by up to 10.8-15.0% on MIMIC-III and 12.6-12.7% on MIMIC-IV for mortality and readmission predictions. In addition to its impressive prediction accuracy, our framework leverages the reasoning capabilities of LLMs, enhancing the trustworthiness of clinical predictions.
One-shot Localization and Segmentation of Medical Images with Foundation Models
Oct 28, 2023



Abstract:Recent advances in Vision Transformers (ViT) and Stable Diffusion (SD) models with their ability to capture rich semantic features of the image have been used for image correspondence tasks on natural images. In this paper, we examine the ability of a variety of pre-trained ViT (DINO, DINOv2, SAM, CLIP) and SD models, trained exclusively on natural images, for solving the correspondence problems on medical images. While many works have made a case for in-domain training, we show that the models trained on natural images can offer good performance on medical images across different modalities (CT,MR,Ultrasound) sourced from various manufacturers, over multiple anatomical regions (brain, thorax, abdomen, extremities), and on wide variety of tasks. Further, we leverage the correspondence with respect to a template image to prompt a Segment Anything (SAM) model to arrive at single shot segmentation, achieving dice range of 62%-90% across tasks, using just one image as reference. We also show that our single-shot method outperforms the recently proposed few-shot segmentation method - UniverSeg (Dice range 47%-80%) on most of the semantic segmentation tasks(six out of seven) across medical imaging modalities.
Improving Early Sepsis Prediction with Multi Modal Learning
Jul 23, 2021
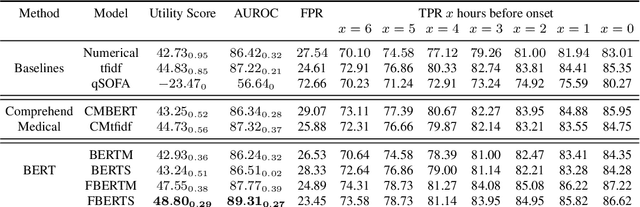


Abstract:Sepsis is a life-threatening disease with high morbidity, mortality and healthcare costs. The early prediction and administration of antibiotics and intravenous fluids is considered crucial for the treatment of sepsis and can save potentially millions of lives and billions in health care costs. Professional clinical care practitioners have proposed clinical criterion which aid in early detection of sepsis; however, performance of these criterion is often limited. Clinical text provides essential information to estimate the severity of the sepsis in addition to structured clinical data. In this study, we explore how clinical text can complement structured data towards early sepsis prediction task. In this paper, we propose multi modal model which incorporates both structured data in the form of patient measurements as well as textual notes on the patient. We employ state-of-the-art NLP models such as BERT and a highly specialized NLP model in Amazon Comprehend Medical to represent the text. On the MIMIC-III dataset containing records of ICU admissions, we show that by using these notes, one achieves an improvement of 6.07 points in a standard utility score for Sepsis prediction and 2.89% in AUROC score. Our methods significantly outperforms a clinical criteria suggested by experts, qSOFA, as well as the winning model of the PhysioNet Computing in Cardiology Challenge for predicting Sepsis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge