Jimeng Sun
Neural Signals Generate Clinical Notes in the Wild
Jan 29, 2026Abstract:Generating clinical reports that summarize abnormal patterns, diagnostic findings, and clinical interpretations from long-term EEG recordings remains labor-intensive. We curate a large-scale clinical EEG dataset with $9{,}922$ reports paired with approximately $11{,}000$ hours of EEG recordings from $9{,}048$ patients. We therefore develop CELM, the first clinical EEG-to-Language foundation model capable of summarizing long-duration, variable-length EEG recordings and performing end-to-end clinical report generation at multiple scales, including recording description, background activity, epileptiform abnormalities, events/seizures, and impressions. Experimental results show that, with patient history supervision, our method achieves $70\%$--$95\%$ average relative improvements in standard generation metrics (e.g., ROUGE-1 and METEOR) from $0.2$--$0.3$ to $0.4$--$0.6$. In the zero-shot setting without patient history, CELM attains generation scores in the range of $0.43$--$0.52$, compared to baselines of $0.17$--$0.26$. CELM integrates pretrained EEG foundation models with language models to enable scalable multimodal learning. We release our model and benchmark construction pipeline at [URL].
PyHealth 2.0: A Comprehensive Open-Source Toolkit for Accessible and Reproducible Clinical Deep Learning
Jan 23, 2026Abstract:Difficulty replicating baselines, high computational costs, and required domain expertise create persistent barriers to clinical AI research. To address these challenges, we introduce PyHealth 2.0, an enhanced clinical deep learning toolkit that enables predictive modeling in as few as 7 lines of code. PyHealth 2.0 offers three key contributions: (1) a comprehensive toolkit addressing reproducibility and compatibility challenges by unifying 15+ datasets, 20+ clinical tasks, 25+ models, 5+ interpretability methods, and uncertainty quantification including conformal prediction within a single framework that supports diverse clinical data modalities - signals, imaging, and electronic health records - with translation of 5+ medical coding standards; (2) accessibility-focused design accommodating multimodal data and diverse computational resources with up to 39x faster processing and 20x lower memory usage, enabling work from 16GB laptops to production systems; and (3) an active open-source community of 400+ members lowering domain expertise barriers through extensive documentation, reproducible research contributions, and collaborations with academic health systems and industry partners, including multi-language support via RHealth. PyHealth 2.0 establishes an open-source foundation and community advancing accessible, reproducible healthcare AI. Available at pip install pyhealth.
$\texttt{AMEND++}$: Benchmarking Eligibility Criteria Amendments in Clinical Trials
Jan 09, 2026Abstract:Clinical trial amendments frequently introduce delays, increased costs, and administrative burden, with eligibility criteria being the most commonly amended component. We introduce \textit{eligibility criteria amendment prediction}, a novel NLP task that aims to forecast whether the eligibility criteria of an initial trial protocol will undergo future amendments. To support this task, we release $\texttt{AMEND++}$, a benchmark suite comprising two datasets: $\texttt{AMEND}$, which captures eligibility-criteria version histories and amendment labels from public clinical trials, and $\verb|AMEND_LLM|$, a refined subset curated using an LLM-based denoising pipeline to isolate substantive changes. We further propose $\textit{Change-Aware Masked Language Modeling}$ (CAMLM), a revision-aware pretraining strategy that leverages historical edits to learn amendment-sensitive representations. Experiments across diverse baselines show that CAMLM consistently improves amendment prediction, enabling more robust and cost-effective clinical trial design.
Adaptation of Agentic AI
Dec 22, 2025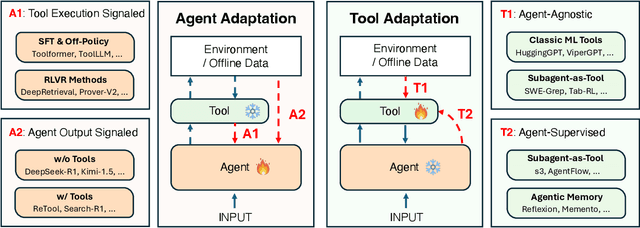
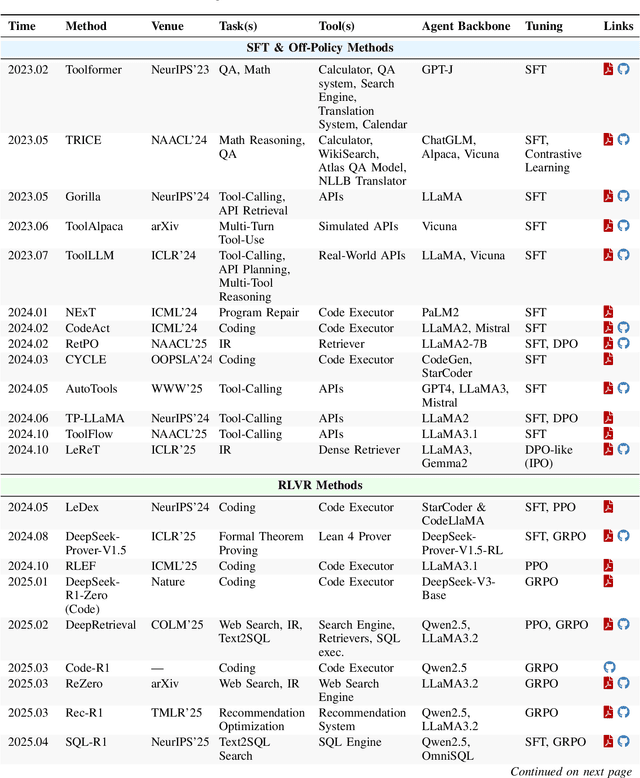
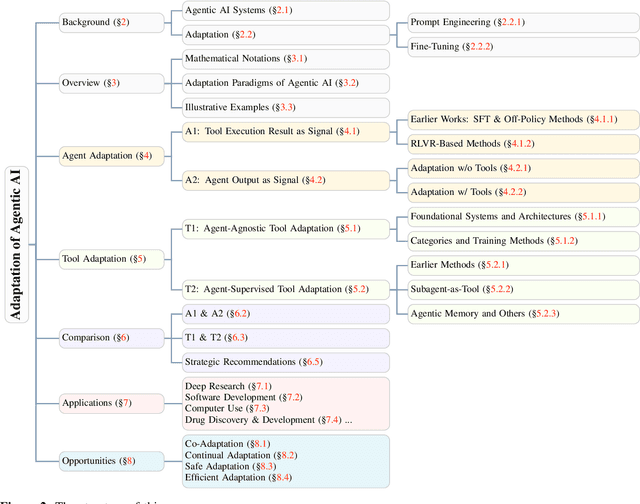
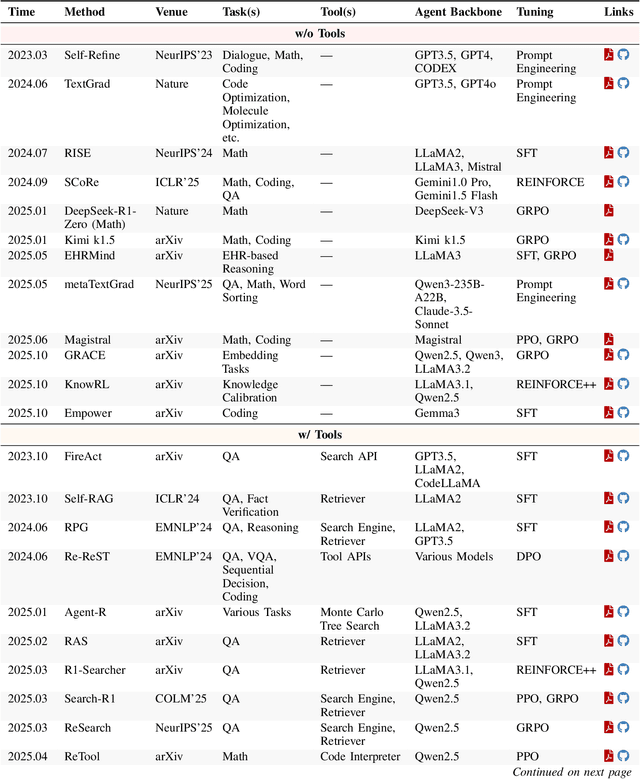
Abstract:Cutting-edge agentic AI systems are built on foundation models that can be adapted to plan, reason, and interact with external tools to perform increasingly complex and specialized tasks. As these systems grow in capability and scope, adaptation becomes a central mechanism for improving performance, reliability, and generalization. In this paper, we unify the rapidly expanding research landscape into a systematic framework that spans both agent adaptations and tool adaptations. We further decompose these into tool-execution-signaled and agent-output-signaled forms of agent adaptation, as well as agent-agnostic and agent-supervised forms of tool adaptation. We demonstrate that this framework helps clarify the design space of adaptation strategies in agentic AI, makes their trade-offs explicit, and provides practical guidance for selecting or switching among strategies during system design. We then review the representative approaches in each category, analyze their strengths and limitations, and highlight key open challenges and future opportunities. Overall, this paper aims to offer a conceptual foundation and practical roadmap for researchers and practitioners seeking to build more capable, efficient, and reliable agentic AI systems.
Prostate-VarBench: A Benchmark with Interpretable TabNet Framework for Prostate Cancer Variant Classification
Nov 12, 2025Abstract:Variants of Uncertain Significance (VUS) limit the clinical utility of prostate cancer genomics by delaying diagnosis and therapy when evidence for pathogenicity or benignity is incomplete. Progress is further limited by inconsistent annotations across sources and the absence of a prostate-specific benchmark for fair comparison. We introduce Prostate-VarBench, a curated pipeline for creating prostate-specific benchmarks that integrates COSMIC (somatic cancer mutations), ClinVar (expert-curated clinical variants), and TCGA-PRAD (prostate tumor genomics from The Cancer Genome Atlas) into a harmonized dataset of 193,278 variants supporting patient- or gene-aware splits to prevent data leakage. To ensure data integrity, we corrected a Variant Effect Predictor (VEP) issue that merged multiple transcript records, introducing ambiguity in clinical significance fields. We then standardized 56 interpretable features across eight clinically relevant tiers, including population frequency, variant type, and clinical context. AlphaMissense pathogenicity scores were incorporated to enhance missense variant classification and reduce VUS uncertainty. Building on this resource, we trained an interpretable TabNet model to classify variant pathogenicity, whose step-wise sparse masks provide per-case rationales consistent with molecular tumor board review practices. On the held-out test set, the model achieved 89.9% accuracy with balanced class metrics, and the VEP correction yields an 6.5% absolute reduction in VUS.
Utilizing Training Data to Improve LLM Reasoning for Tabular Understanding
Aug 26, 2025Abstract:Automated tabular understanding and reasoning are essential tasks for data scientists. Recently, Large language models (LLMs) have become increasingly prevalent in tabular reasoning tasks. Previous work focuses on (1) finetuning LLMs using labeled data or (2) Training-free prompting LLM agents using chain-of-thought (CoT). Finetuning offers dataset-specific learning at the cost of generalizability. Training-free prompting is highly generalizable but does not take full advantage of training data. In this paper, we propose a novel prompting-based reasoning approach, Learn then Retrieve: LRTab, which integrates the benefits of both by retrieving relevant information learned from training data. We first use prompting to obtain CoT responses over the training data. For incorrect CoTs, we prompt the LLM to predict Prompt Conditions to avoid the error, learning insights from the data. We validate the effectiveness of Prompt Conditions using validation data. Finally, at inference time, we retrieve the most relevant Prompt Conditions for additional context for table understanding. We provide comprehensive experiments on WikiTQ and Tabfact, showing that LRTab is interpretable, cost-efficient, and can outperform previous baselines in tabular reasoning.
CAPO: Reinforcing Consistent Reasoning in Medical Decision-Making
Jun 15, 2025
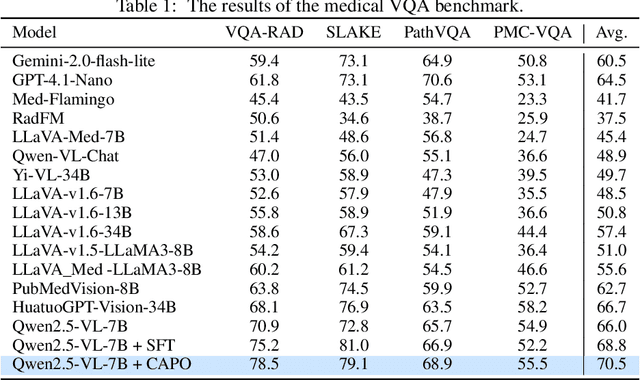
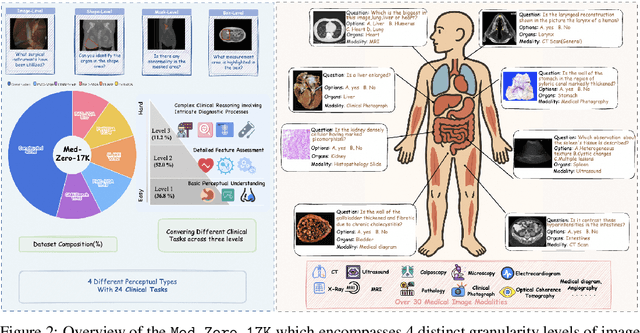
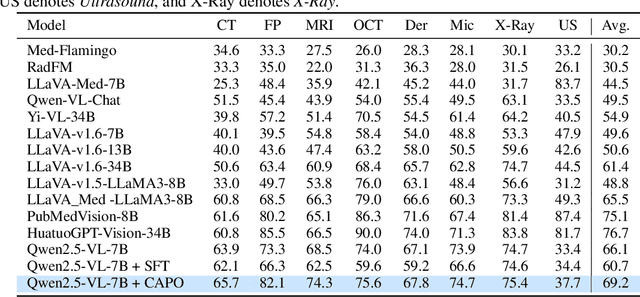
Abstract:In medical visual question answering (Med-VQA), achieving accurate responses relies on three critical steps: precise perception of medical imaging data, logical reasoning grounded in visual input and textual questions, and coherent answer derivation from the reasoning process. Recent advances in general vision-language models (VLMs) show that large-scale reinforcement learning (RL) could significantly enhance both reasoning capabilities and overall model performance. However, their application in medical domains is hindered by two fundamental challenges: 1) misalignment between perceptual understanding and reasoning stages, and 2) inconsistency between reasoning pathways and answer generation, both compounded by the scarcity of high-quality medical datasets for effective large-scale RL. In this paper, we first introduce Med-Zero-17K, a curated dataset for pure RL-based training, encompassing over 30 medical image modalities and 24 clinical tasks. Moreover, we propose a novel large-scale RL framework for Med-VLMs, Consistency-Aware Preference Optimization (CAPO), which integrates rewards to ensure fidelity between perception and reasoning, consistency in reasoning-to-answer derivation, and rule-based accuracy for final responses. Extensive experiments on both in-domain and out-of-domain scenarios demonstrate the superiority of our method over strong VLM baselines, showcasing strong generalization capability to 3D Med-VQA benchmarks and R1-like training paradigms.
Training LLMs for EHR-Based Reasoning Tasks via Reinforcement Learning
May 30, 2025Abstract:We present EHRMIND, a practical recipe for adapting large language models (LLMs) to complex clinical reasoning tasks using reinforcement learning with verifiable rewards (RLVR). While RLVR has succeeded in mathematics and coding, its application to healthcare contexts presents unique challenges due to the specialized knowledge and reasoning required for electronic health record (EHR) interpretation. Our pilot study on the MEDCALC benchmark reveals two key failure modes: (1) misapplied knowledge, where models possess relevant medical knowledge but apply it incorrectly, and (2) missing knowledge, where models lack essential domain knowledge. To address these cases, EHRMIND applies a two-stage solution: a lightweight supervised fine-tuning (SFT) warm-up that injects missing domain knowledge, stabilizes subsequent training, and encourages structured, interpretable outputs; followed by RLVR, which reinforces outcome correctness and refines the model's decision-making. We demonstrate the effectiveness of our method across diverse clinical applications, including medical calculations (MEDCALC), patient-trial matching (TREC CLINICAL TRIALS), and disease diagnosis (EHRSHOT). EHRMIND delivers consistent gains in accuracy, interpretability, and cross-task generalization. These findings offer practical guidance for applying RLVR to enhance LLM capabilities in healthcare settings.
Reinforcement Learning for Out-of-Distribution Reasoning in LLMs: An Empirical Study on Diagnosis-Related Group Coding
May 28, 2025Abstract:Diagnosis-Related Group (DRG) codes are essential for hospital reimbursement and operations but require labor-intensive assignment. Large Language Models (LLMs) struggle with DRG coding due to the out-of-distribution (OOD) nature of the task: pretraining corpora rarely contain private clinical or billing data. We introduce DRG-Sapphire, which uses large-scale reinforcement learning (RL) for automated DRG coding from clinical notes. Built on Qwen2.5-7B and trained with Group Relative Policy Optimization (GRPO) using rule-based rewards, DRG-Sapphire introduces a series of RL enhancements to address domain-specific challenges not seen in previous mathematical tasks. Our model achieves state-of-the-art accuracy on the MIMIC-IV benchmark and generates physician-validated reasoning for DRG assignments, significantly enhancing explainability. Our study further sheds light on broader challenges of applying RL to knowledge-intensive, OOD tasks. We observe that RL performance scales approximately linearly with the logarithm of the number of supervised fine-tuning (SFT) examples, suggesting that RL effectiveness is fundamentally constrained by the domain knowledge encoded in the base model. For OOD tasks like DRG coding, strong RL performance requires sufficient knowledge infusion prior to RL. Consequently, scaling SFT may be more effective and computationally efficient than scaling RL alone for such tasks.
BioDSA-1K: Benchmarking Data Science Agents for Biomedical Research
May 22, 2025Abstract:Validating scientific hypotheses is a central challenge in biomedical research, and remains difficult for artificial intelligence (AI) agents due to the complexity of real-world data analysis and evidence interpretation. In this work, we present BioDSA-1K, a benchmark designed to evaluate AI agents on realistic, data-driven biomedical hypothesis validation tasks. BioDSA-1K consists of 1,029 hypothesis-centric tasks paired with 1,177 analysis plans, curated from over 300 published biomedical studies to reflect the structure and reasoning found in authentic research workflows. Each task includes a structured hypothesis derived from the original study's conclusions, expressed in the affirmative to reflect the language of scientific reporting, and one or more pieces of supporting evidence grounded in empirical data tables. While these hypotheses mirror published claims, they remain testable using standard statistical or machine learning methods. The benchmark enables evaluation along four axes: (1) hypothesis decision accuracy, (2) alignment between evidence and conclusion, (3) correctness of the reasoning process, and (4) executability of the AI-generated analysis code. Importantly, BioDSA-1K includes non-verifiable hypotheses: cases where the available data are insufficient to support or refute a claim, reflecting a common yet underexplored scenario in real-world science. We propose BioDSA-1K as a foundation for building and evaluating generalizable, trustworthy AI agents for biomedical discovery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge