Zheng Chen
LSGQuant: Layer-Sensitivity Guided Quantization for One-Step Diffusion Real-World Video Super-Resolution
Feb 03, 2026Abstract:One-Step Diffusion Models have demonstrated promising capability and fast inference in video super-resolution (VSR) for real-world. Nevertheless, the substantial model size and high computational cost of Diffusion Transformers (DiTs) limit downstream applications. While low-bit quantization is a common approach for model compression, the effectiveness of quantized models is challenged by the high dynamic range of input latent and diverse layer behaviors. To deal with these challenges, we introduce LSGQuant, a layer-sensitivity guided quantizing approach for one-step diffusion-based real-world VSR. Our method incorporates a Dynamic Range Adaptive Quantizer (DRAQ) to fit video token activations. Furthermore, we estimate layer sensitivity and implement a Variance-Oriented Layer Training Strategy (VOLTS) by analyzing layer-wise statistics in calibration. We also introduce Quantization-Aware Optimization (QAO) to jointly refine the quantized branch and a retained high-precision branch. Extensive experiments demonstrate that our method has nearly performance to origin model with full-precision and significantly exceeds existing quantization techniques. Code is available at: https://github.com/zhengchen1999/LSGQuant.
BinaryDemoire: Moiré-Aware Binarization for Image Demoiréing
Feb 03, 2026Abstract:Image demoiréing aims to remove structured moiré artifacts in recaptured imagery, where degradations are highly frequency-dependent and vary across scales and directions. While recent deep networks achieve high-quality restoration, their full-precision designs remain costly for deployment. Binarization offers an extreme compression regime by quantizing both activations and weights to 1-bit. Yet, it has been rarely studied for demoiréing and performs poorly when naively applied. In this work, we propose BinaryDemoire, a binarized demoiréing framework that explicitly accommodates the frequency structure of moiré degradations. First, we introduce a moiré-aware binary gate (MABG) that extracts lightweight frequency descriptors together with activation statistics. It predicts channel-wise gating coefficients to condition the aggregation of binary convolution responses. Second, we design a shuffle-grouped residual adapter (SGRA) that performs structured sparse shortcut alignment. It further integrates interleaved mixing to promote information exchange across different channel partitions. Extensive experiments on four benchmarks demonstrate that the proposed BinaryDemoire surpasses current binarization methods. Code: https://github.com/zhengchen1999/BinaryDemoire.
VEQ: Modality-Adaptive Quantization for MoE Vision-Language Models
Feb 01, 2026Abstract:Mixture-of-Experts(MoE) Vision-Language Models (VLMs) offer remarkable performance but incur prohibitive memory and computational costs, making compression essential. Post-Training Quantization (PTQ) is an effective training-free technique to address the massive memory and computation overhead. Existing quantization paradigms fall short as they are oblivious to two critical forms of heterogeneity: the inherent discrepancy between vision and language tokens, and the non-uniform contribution of different experts. To bridge this gap, we propose Visual Expert Quantization (VEQ), a dual-aware quantization framework designed to simultaneously accommodate cross-modal differences and heterogeneity between experts. Specifically, VEQ incorporates 1)Modality-expert-aware Quantization, which utilizes expert activation frequency to prioritize error minimization for pivotal experts, and 2)Modality-affinity-aware Quantization, which constructs an enhanced Hessian matrix by integrating token-expert affinity with modality information to guide the calibration process. Extensive experiments across diverse benchmarks verify that VEQ consistently outperforms state-of-the-art baselines. Specifically, under the W3A16 configuration, our method achieves significant average accuracy gains of 2.04\% on Kimi-VL and 3.09\% on Qwen3-VL compared to the previous SOTA quantization methods, demonstrating superior robustness across various multimodal tasks. Our code will be available at https://github.com/guangshuoqin/VEQ.
Neural Signals Generate Clinical Notes in the Wild
Jan 29, 2026Abstract:Generating clinical reports that summarize abnormal patterns, diagnostic findings, and clinical interpretations from long-term EEG recordings remains labor-intensive. We curate a large-scale clinical EEG dataset with $9{,}922$ reports paired with approximately $11{,}000$ hours of EEG recordings from $9{,}048$ patients. We therefore develop CELM, the first clinical EEG-to-Language foundation model capable of summarizing long-duration, variable-length EEG recordings and performing end-to-end clinical report generation at multiple scales, including recording description, background activity, epileptiform abnormalities, events/seizures, and impressions. Experimental results show that, with patient history supervision, our method achieves $70\%$--$95\%$ average relative improvements in standard generation metrics (e.g., ROUGE-1 and METEOR) from $0.2$--$0.3$ to $0.4$--$0.6$. In the zero-shot setting without patient history, CELM attains generation scores in the range of $0.43$--$0.52$, compared to baselines of $0.17$--$0.26$. CELM integrates pretrained EEG foundation models with language models to enable scalable multimodal learning. We release our model and benchmark construction pipeline at [URL].
SkyReels-V3 Technique Report
Jan 24, 2026Abstract:Video generation serves as a cornerstone for building world models, where multimodal contextual inference stands as the defining test of capability. In this end, we present SkyReels-V3, a conditional video generation model, built upon a unified multimodal in-context learning framework with diffusion Transformers. SkyReels-V3 model supports three core generative paradigms within a single architecture: reference images-to-video synthesis, video-to-video extension and audio-guided video generation. (i) reference images-to-video model is designed to produce high-fidelity videos with strong subject identity preservation, temporal coherence, and narrative consistency. To enhance reference adherence and compositional stability, we design a comprehensive data processing pipeline that leverages cross frame pairing, image editing, and semantic rewriting, effectively mitigating copy paste artifacts. During training, an image video hybrid strategy combined with multi-resolution joint optimization is employed to improve generalization and robustness across diverse scenarios. (ii) video extension model integrates spatio-temporal consistency modeling with large-scale video understanding, enabling both seamless single-shot continuation and intelligent multi-shot switching with professional cinematographic patterns. (iii) Talking avatar model supports minute-level audio-conditioned video generation by training first-and-last frame insertion patterns and reconstructing key-frame inference paradigms. On the basis of ensuring visual quality, synchronization of audio and videos has been optimized. Extensive evaluations demonstrate that SkyReels-V3 achieves state-of-the-art or near state-of-the-art performance on key metrics including visual quality, instruction following, and specific aspect metrics, approaching leading closed-source systems. Github: https://github.com/SkyworkAI/SkyReels-V3.
A Unified Convergence Analysis for Semi-Decentralized Learning: Sampled-to-Sampled vs. Sampled-to-All Communication
Nov 17, 2025Abstract:In semi-decentralized federated learning, devices primarily rely on device-to-device communication but occasionally interact with a central server. Periodically, a sampled subset of devices uploads their local models to the server, which computes an aggregate model. The server can then either (i) share this aggregate model only with the sampled clients (sampled-to-sampled, S2S) or (ii) broadcast it to all clients (sampled-to-all, S2A). Despite their practical significance, a rigorous theoretical and empirical comparison of these two strategies remains absent. We address this gap by analyzing S2S and S2A within a unified convergence framework that accounts for key system parameters: sampling rate, server aggregation frequency, and network connectivity. Our results, both analytical and experimental, reveal distinct regimes where one strategy outperforms the other, depending primarily on the degree of data heterogeneity across devices. These insights lead to concrete design guidelines for practical semi-decentralized FL deployments.
Navigating the Wild: Pareto-Optimal Visual Decision-Making in Image Space
Nov 11, 2025Abstract:Navigating complex real-world environments requires semantic understanding and adaptive decision-making. Traditional reactive methods without maps often fail in cluttered settings, map-based approaches demand heavy mapping effort, and learning-based solutions rely on large datasets with limited generalization. To address these challenges, we present Pareto-Optimal Visual Navigation, a lightweight image-space framework that combines data-driven semantics, Pareto-optimal decision-making, and visual servoing for real-time navigation.
FideDiff: Efficient Diffusion Model for High-Fidelity Image Motion Deblurring
Oct 02, 2025

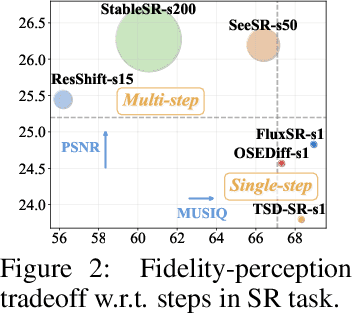

Abstract:Recent advancements in image motion deblurring, driven by CNNs and transformers, have made significant progress. Large-scale pre-trained diffusion models, which are rich in true-world modeling, have shown great promise for high-quality image restoration tasks such as deblurring, demonstrating stronger generative capabilities than CNN and transformer-based methods. However, challenges such as unbearable inference time and compromised fidelity still limit the full potential of the diffusion models. To address this, we introduce FideDiff, a novel single-step diffusion model designed for high-fidelity deblurring. We reformulate motion deblurring as a diffusion-like process where each timestep represents a progressively blurred image, and we train a consistency model that aligns all timesteps to the same clean image. By reconstructing training data with matched blur trajectories, the model learns temporal consistency, enabling accurate one-step deblurring. We further enhance model performance by integrating Kernel ControlNet for blur kernel estimation and introducing adaptive timestep prediction. Our model achieves superior performance on full-reference metrics, surpassing previous diffusion-based methods and matching the performance of other state-of-the-art models. FideDiff offers a new direction for applying pre-trained diffusion models to high-fidelity image restoration tasks, establishing a robust baseline for further advancing diffusion models in real-world industrial applications. Our dataset and code will be available at https://github.com/xyLiu339/FideDiff.
EvoBrain: Dynamic Multi-channel EEG Graph Modeling for Time-evolving Brain Network
Sep 19, 2025Abstract:Dynamic GNNs, which integrate temporal and spatial features in Electroencephalography (EEG) data, have shown great potential in automating seizure detection. However, fully capturing the underlying dynamics necessary to represent brain states, such as seizure and non-seizure, remains a non-trivial task and presents two fundamental challenges. First, most existing dynamic GNN methods are built on temporally fixed static graphs, which fail to reflect the evolving nature of brain connectivity during seizure progression. Second, current efforts to jointly model temporal signals and graph structures and, more importantly, their interactions remain nascent, often resulting in inconsistent performance. To address these challenges, we present the first theoretical analysis of these two problems, demonstrating the effectiveness and necessity of explicit dynamic modeling and time-then-graph dynamic GNN method. Building on these insights, we propose EvoBrain, a novel seizure detection model that integrates a two-stream Mamba architecture with a GCN enhanced by Laplacian Positional Encoding, following neurological insights. Moreover, EvoBrain incorporates explicitly dynamic graph structures, allowing both nodes and edges to evolve over time. Our contributions include (a) a theoretical analysis proving the expressivity advantage of explicit dynamic modeling and time-then-graph over other approaches, (b) a novel and efficient model that significantly improves AUROC by 23% and F1 score by 30%, compared with the dynamic GNN baseline, and (c) broad evaluations of our method on the challenging early seizure prediction tasks.
Steering One-Step Diffusion Model with Fidelity-Rich Decoder for Fast Image Compression
Aug 07, 2025Abstract:Diffusion-based image compression has demonstrated impressive perceptual performance. However, it suffers from two critical drawbacks: (1) excessive decoding latency due to multi-step sampling, and (2) poor fidelity resulting from over-reliance on generative priors. To address these issues, we propose SODEC, a novel single-step diffusion image compression model. We argue that in image compression, a sufficiently informative latent renders multi-step refinement unnecessary. Based on this insight, we leverage a pre-trained VAE-based model to produce latents with rich information, and replace the iterative denoising process with a single-step decoding. Meanwhile, to improve fidelity, we introduce the fidelity guidance module, encouraging output that is faithful to the original image. Furthermore, we design the rate annealing training strategy to enable effective training under extremely low bitrates. Extensive experiments show that SODEC significantly outperforms existing methods, achieving superior rate-distortion-perception performance. Moreover, compared to previous diffusion-based compression models, SODEC improves decoding speed by more than 20$\times$. Code is released at: https://github.com/zhengchen1999/SODEC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge