Hanyin Wang
Reinforcement Learning for Out-of-Distribution Reasoning in LLMs: An Empirical Study on Diagnosis-Related Group Coding
May 28, 2025Abstract:Diagnosis-Related Group (DRG) codes are essential for hospital reimbursement and operations but require labor-intensive assignment. Large Language Models (LLMs) struggle with DRG coding due to the out-of-distribution (OOD) nature of the task: pretraining corpora rarely contain private clinical or billing data. We introduce DRG-Sapphire, which uses large-scale reinforcement learning (RL) for automated DRG coding from clinical notes. Built on Qwen2.5-7B and trained with Group Relative Policy Optimization (GRPO) using rule-based rewards, DRG-Sapphire introduces a series of RL enhancements to address domain-specific challenges not seen in previous mathematical tasks. Our model achieves state-of-the-art accuracy on the MIMIC-IV benchmark and generates physician-validated reasoning for DRG assignments, significantly enhancing explainability. Our study further sheds light on broader challenges of applying RL to knowledge-intensive, OOD tasks. We observe that RL performance scales approximately linearly with the logarithm of the number of supervised fine-tuning (SFT) examples, suggesting that RL effectiveness is fundamentally constrained by the domain knowledge encoded in the base model. For OOD tasks like DRG coding, strong RL performance requires sufficient knowledge infusion prior to RL. Consequently, scaling SFT may be more effective and computationally efficient than scaling RL alone for such tasks.
Process-Supervised Reward Models for Clinical Note Generation: A Scalable Approach Guided by Domain Expertise
Dec 17, 2024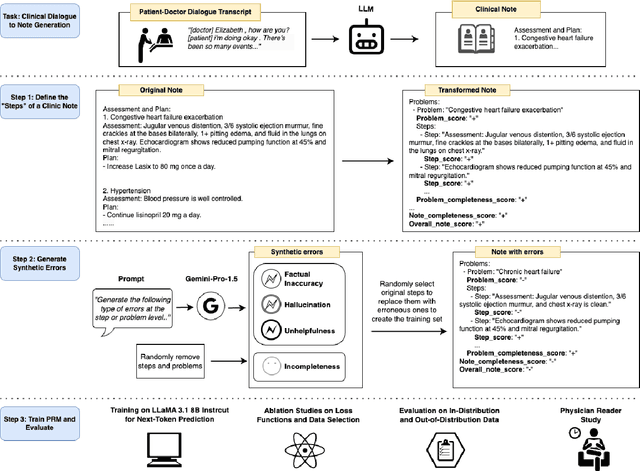
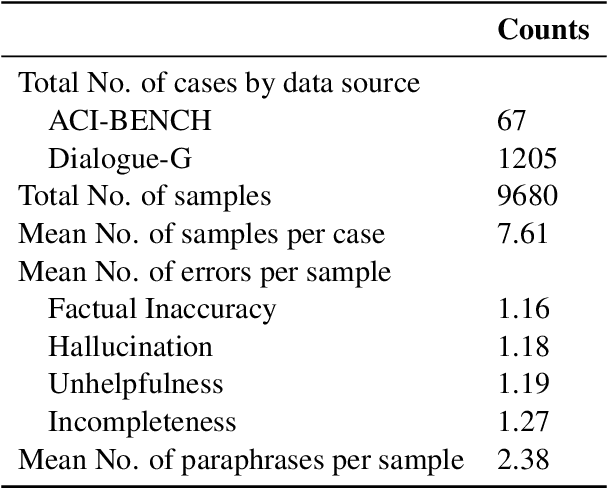
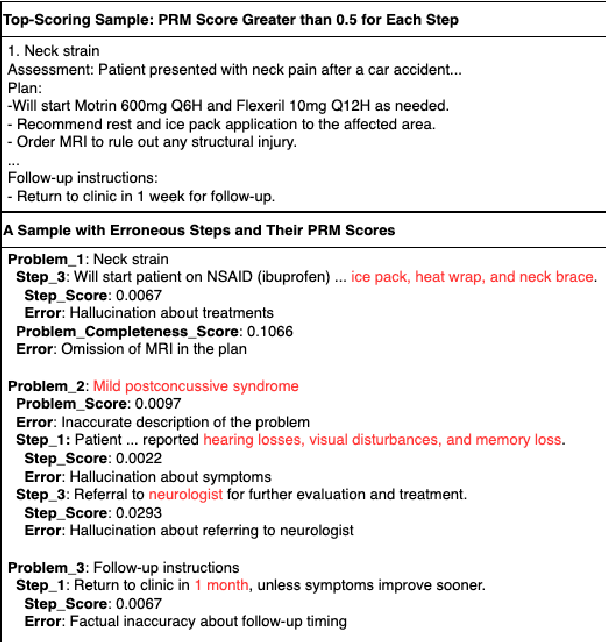
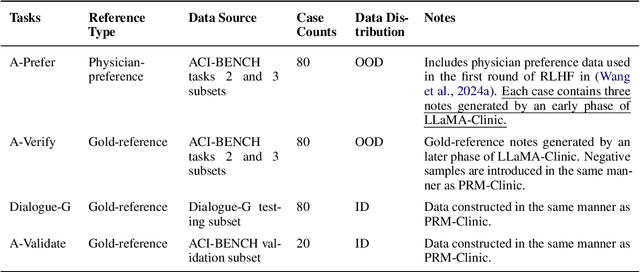
Abstract:Process-supervised reward models (PRMs), which verify large language model (LLM) outputs step-by-step, have achieved significant success in mathematical and coding problems. However, their application to other domains remains largely unexplored. In this work, we train a PRM to provide step-level reward signals for clinical notes generated by LLMs from patient-doctor dialogues. Guided by real-world clinician expertise, we carefully designed step definitions for clinical notes and utilized Gemini-Pro 1.5 to automatically generate process supervision data at scale. Our proposed PRM, trained on the LLaMA-3.1 8B instruct model, demonstrated superior performance compared to Gemini-Pro 1.5 and an outcome-supervised reward model (ORM) across two key evaluations: (1) the accuracy of selecting gold-reference samples from error-containing samples, achieving 98.8% (versus 61.3% for ORM and 93.8% for Gemini-Pro 1.5), and (2) the accuracy of selecting physician-preferred notes, achieving 56.2% (compared to 51.2% for ORM and 50.0% for Gemini-Pro 1.5). Additionally, we conducted ablation studies to determine optimal loss functions and data selection strategies, along with physician reader studies to explore predictors of downstream Best-of-N performance. Our promising results suggest the potential of PRMs to extend beyond the clinical domain, offering a scalable and effective solution for diverse generative tasks.
A Perspective for Adapting Generalist AI to Specialized Medical AI Applications and Their Challenges
Oct 28, 2024
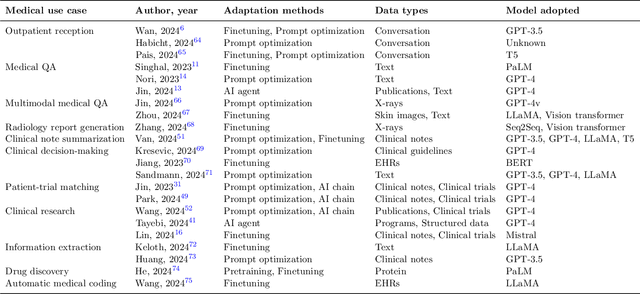

Abstract:The integration of Large Language Models (LLMs) into medical applications has sparked widespread interest across the healthcare industry, from drug discovery and development to clinical decision support, assisting telemedicine, medical devices, and healthcare insurance applications. This perspective paper aims to discuss the inner workings of building LLM-powered medical AI applications and introduces a comprehensive framework for their development. We review existing literature and outline the unique challenges of applying LLMs in specialized medical contexts. Additionally, we introduce a three-step framework to organize medical LLM research activities: 1) Modeling: breaking down complex medical workflows into manageable steps for developing medical-specific models; 2) Optimization: optimizing the model performance with crafted prompts and integrating external knowledge and tools, and 3) System engineering: decomposing complex tasks into subtasks and leveraging human expertise for building medical AI applications. Furthermore, we offer a detailed use case playbook that describes various LLM-powered medical AI applications, such as optimizing clinical trial design, enhancing clinical decision support, and advancing medical imaging analysis. Finally, we discuss various challenges and considerations for building medical AI applications with LLMs, such as handling hallucination issues, data ownership and compliance, privacy, intellectual property considerations, compute cost, sustainability issues, and responsible AI requirements.
Towards Adapting Open-Source Large Language Models for Expert-Level Clinical Note Generation
Apr 25, 2024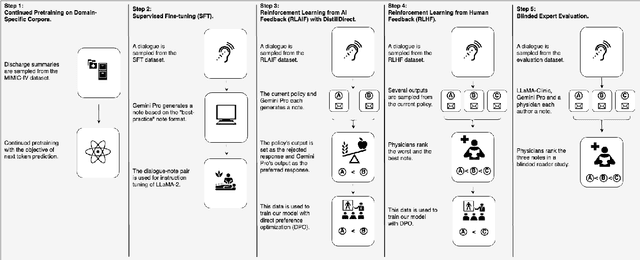
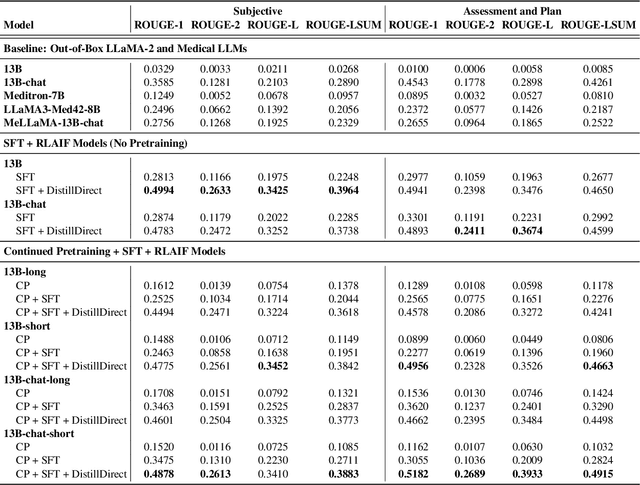
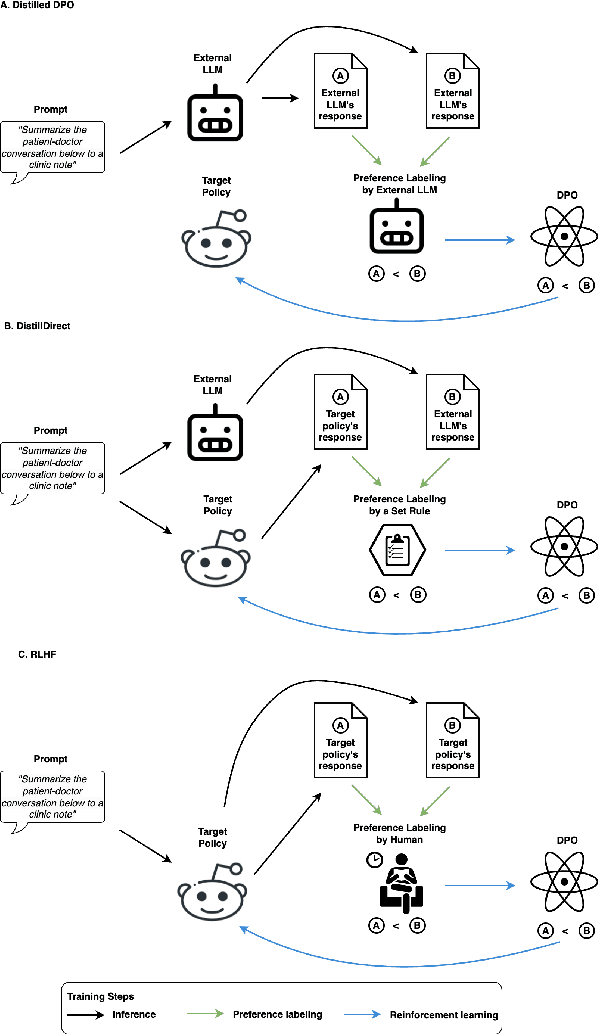
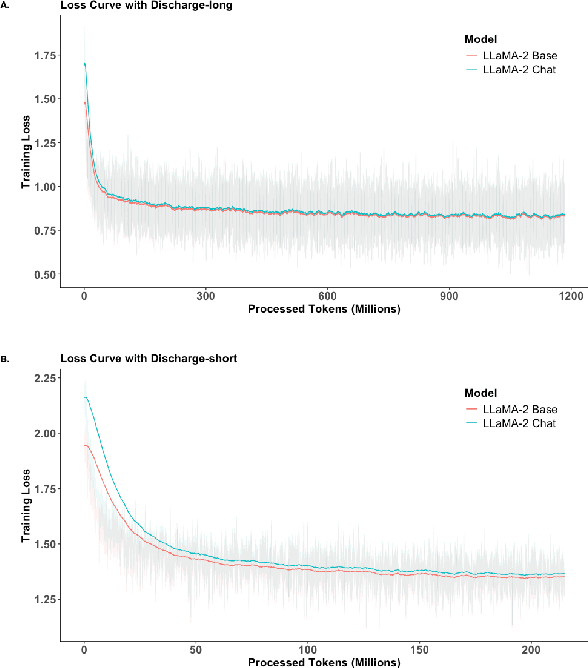
Abstract:Large Language Models (LLMs) have shown promising capabilities in handling clinical text summarization tasks. In this study, we demonstrate that a small open-source LLM can be effectively trained to generate high-quality clinical notes from outpatient patient-doctor dialogues. We achieve this through a comprehensive domain- and task-specific adaptation process for the LLaMA-2 13 billion parameter model. This process incorporates continued pre-training, supervised fine-tuning, and reinforcement learning from both AI and human feedback. We introduced an enhanced approach, termed DistillDirect, for performing on-policy reinforcement learning with Gemini Pro serving as the teacher model. Our resulting model, LLaMA-Clinic, is capable of generating clinical notes that are comparable in quality to those authored by physicians. In a blinded physician reader study, the majority (90.4%) of individual evaluations rated the notes generated by LLaMA-Clinic as "acceptable" or higher across all three criteria: real-world readiness, completeness, and accuracy. Notably, in the more challenging "Assessment and Plan" section, LLaMA-Clinic scored higher (4.2/5) in real-world readiness compared to physician-authored notes (4.1/5). Additionally, we identified caveats in public clinical note datasets, such as ACI-BENCH. We highlight key considerations for future clinical note-generation tasks, emphasizing the importance of pre-defining a best-practice note format. Overall, our research demonstrates the potential and feasibility of training smaller, open-source LLMs to assist with clinical documentation, capitalizing on healthcare institutions' access to patient records and domain expertise. We have made our newly created synthetic clinic dialogue-note dataset and the physician feedback dataset publicly available to foster future research in this field.
DRG-LLaMA : Tuning LLaMA Model to Predict Diagnosis-related Group for Hospitalized Patients
Sep 29, 2023



Abstract:In the U.S. inpatient payment system, the Diagnosis-Related Group (DRG) is pivotal, but its assignment process is inefficient. The study introduces DRG-LLaMA, an advanced large language model (LLM) fine-tuned on clinical notes to enhance DRGs assignment. Utilizing LLaMA as the foundational model and optimizing it through Low-Rank Adaptation (LoRA) on 236,192 MIMIC-IV discharge summaries, our DRG-LLaMA-7B model exhibited a noteworthy macro-averaged F1 score of 0.327, a top-1 prediction accuracy of 52.0%, and a macro-averaged Area Under the Curve (AUC) of 0.986, with a maximum input token length of 512. This model surpassed the performance of prior leading models in DRG prediction, showing a relative improvement of 40.3% and 35.7% in macro-averaged F1 score compared to ClinicalBERT and CAML, respectively. Applied to base DRG and complication or comorbidity (CC)/major complication or comorbidity (MCC) prediction, DRG-LLaMA achieved a top-1 prediction accuracy of 67.8% and 67.5%, respectively. Additionally, our findings indicate that DRG-LLaMA's performance correlates with increased model parameters and input context lengths.
Deep Reinforcement Learning for Efficient and Fair Allocation of Health Care Resources
Sep 15, 2023



Abstract:Scarcity of health care resources could result in the unavoidable consequence of rationing. For example, ventilators are often limited in supply, especially during public health emergencies or in resource-constrained health care settings, such as amid the pandemic of COVID-19. Currently, there is no universally accepted standard for health care resource allocation protocols, resulting in different governments prioritizing patients based on various criteria and heuristic-based protocols. In this study, we investigate the use of reinforcement learning for critical care resource allocation policy optimization to fairly and effectively ration resources. We propose a transformer-based deep Q-network to integrate the disease progression of individual patients and the interaction effects among patients during the critical care resource allocation. We aim to improve both fairness of allocation and overall patient outcomes. Our experiments demonstrate that our method significantly reduces excess deaths and achieves a more equitable distribution under different levels of ventilator shortage, when compared to existing severity-based and comorbidity-based methods in use by different governments. Our source code is included in the supplement and will be released on Github upon publication.
A Comparative Study of Pretrained Language Models for Long Clinical Text
Jan 27, 2023



Abstract:Objective: Clinical knowledge enriched transformer models (e.g., ClinicalBERT) have state-of-the-art results on clinical NLP (natural language processing) tasks. One of the core limitations of these transformer models is the substantial memory consumption due to their full self-attention mechanism, which leads to the performance degradation in long clinical texts. To overcome this, we propose to leverage long-sequence transformer models (e.g., Longformer and BigBird), which extend the maximum input sequence length from 512 to 4096, to enhance the ability to model long-term dependencies in long clinical texts. Materials and Methods: Inspired by the success of long sequence transformer models and the fact that clinical notes are mostly long, we introduce two domain enriched language models, Clinical-Longformer and Clinical-BigBird, which are pre-trained on a large-scale clinical corpus. We evaluate both language models using 10 baseline tasks including named entity recognition, question answering, natural language inference, and document classification tasks. Results: The results demonstrate that Clinical-Longformer and Clinical-BigBird consistently and significantly outperform ClinicalBERT and other short-sequence transformers in all 10 downstream tasks and achieve new state-of-the-art results. Discussion: Our pre-trained language models provide the bedrock for clinical NLP using long texts. We have made our source code available at https://github.com/luoyuanlab/Clinical-Longformer, and the pre-trained models available for public download at: https://huggingface.co/yikuan8/Clinical-Longformer. Conclusion: This study demonstrates that clinical knowledge enriched long-sequence transformers are able to learn long-term dependencies in long clinical text. Our methods can also inspire the development of other domain-enriched long-sequence transformers.
Multimodal Machine Learning in Precision Health
Apr 10, 2022



Abstract:As machine learning and artificial intelligence are more frequently being leveraged to tackle problems in the health sector, there has been increased interest in utilizing them in clinical decision-support. This has historically been the case in single modal data such as electronic health record data. Attempts to improve prediction and resemble the multimodal nature of clinical expert decision-making this has been met in the computational field of machine learning by a fusion of disparate data. This review was conducted to summarize this field and identify topics ripe for future research. We conducted this review in accordance with the PRISMA (Preferred Reporting Items for Systematic reviews and Meta-Analyses) extension for Scoping Reviews to characterize multi-modal data fusion in health. We used a combination of content analysis and literature searches to establish search strings and databases of PubMed, Google Scholar, and IEEEXplore from 2011 to 2021. A final set of 125 articles were included in the analysis. The most common health areas utilizing multi-modal methods were neurology and oncology. However, there exist a wide breadth of current applications. The most common form of information fusion was early fusion. Notably, there was an improvement in predictive performance performing heterogeneous data fusion. Lacking from the papers were clear clinical deployment strategies and pursuit of FDA-approved tools. These findings provide a map of the current literature on multimodal data fusion as applied to health diagnosis/prognosis problems. Multi-modal machine learning, while more robust in its estimations over unimodal methods, has drawbacks in its scalability and the time-consuming nature of information concatenation.
Clinical-Longformer and Clinical-BigBird: Transformers for long clinical sequences
Feb 12, 2022



Abstract:Transformers-based models, such as BERT, have dramatically improved the performance for various natural language processing tasks. The clinical knowledge enriched model, namely ClinicalBERT, also achieved state-of-the-art results when performed on clinical named entity recognition and natural language inference tasks. One of the core limitations of these transformers is the substantial memory consumption due to their full self-attention mechanism. To overcome this, long sequence transformer models, e.g. Longformer and BigBird, were proposed with the idea of sparse attention mechanism to reduce the memory usage from quadratic to the sequence length to a linear scale. These models extended the maximum input sequence length from 512 to 4096, which enhanced the ability of modeling long-term dependency and consequently achieved optimal results in a variety of tasks. Inspired by the success of these long sequence transformer models, we introduce two domain enriched language models, namely Clinical-Longformer and Clinical-BigBird, which are pre-trained from large-scale clinical corpora. We evaluate both pre-trained models using 10 baseline tasks including named entity recognition, question answering, and document classification tasks. The results demonstrate that Clinical-Longformer and Clinical-BigBird consistently and significantly outperform ClinicalBERT as well as other short-sequence transformers in all downstream tasks. We have made our source code available at [https://github.com/luoyuanlab/Clinical-Longformer] the pre-trained models available for public download at: [https://huggingface.co/yikuan8/Clinical-Longformer].
Disparities in Social Determinants among Performances of Mortality Prediction with Machine Learning for Sepsis Patients
Dec 15, 2021



Abstract:Background Sepsis is one of the most life-threatening circumstances for critically ill patients in the US, while a standardized criteria for sepsis identification is still under development. Disparities in social determinants of sepsis patients can interfere with the risk prediction performances using machine learning. Methods Disparities in social determinants, including race, gender, marital status, insurance types and languages, among patients identified by six available sepsis criteria were revealed by forest plots. Sixteen machine learning classifiers were trained to predict in-hospital mortality for sepsis patients. The performance of the trained model was tested on the entire randomly conducted test set and each sub-population built based on each of the following social determinants: race, gender, marital status, insurance type, and language. Results We analyzed a total of 11,791 critical care patients from the MIMIC-III database. Within the population identified by each sepsis identification method, significant differences were observed among sub-populations regarding race, marital status, insurance type, and language. On the 5,783 sepsis patients identified by the Sepsis-3 criteria statistically significant performance decreases for mortality prediction were observed when applying the trained machine learning model on Asian and Hispanic patients. With pairwise comparison, we detected performance discrepancies in mortality prediction between Asian and White patients, Asians and patients of other races, as well as English-speaking and Spanish-speaking patients. Conclusions Disparities in proportions of patients identified by various sepsis criteria were detected among the different social determinant groups. To achieve accurate diagnosis, a versatile diagnostic system for sepsis is needed to overcome the social determinant disparities of patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge