Yikuan Li
Deep Reinforcement Learning for Efficient and Fair Allocation of Health Care Resources
Sep 15, 2023



Abstract:Scarcity of health care resources could result in the unavoidable consequence of rationing. For example, ventilators are often limited in supply, especially during public health emergencies or in resource-constrained health care settings, such as amid the pandemic of COVID-19. Currently, there is no universally accepted standard for health care resource allocation protocols, resulting in different governments prioritizing patients based on various criteria and heuristic-based protocols. In this study, we investigate the use of reinforcement learning for critical care resource allocation policy optimization to fairly and effectively ration resources. We propose a transformer-based deep Q-network to integrate the disease progression of individual patients and the interaction effects among patients during the critical care resource allocation. We aim to improve both fairness of allocation and overall patient outcomes. Our experiments demonstrate that our method significantly reduces excess deaths and achieves a more equitable distribution under different levels of ventilator shortage, when compared to existing severity-based and comorbidity-based methods in use by different governments. Our source code is included in the supplement and will be released on Github upon publication.
Deep Reinforcement Learning for Cost-Effective Medical Diagnosis
Feb 28, 2023Abstract:Dynamic diagnosis is desirable when medical tests are costly or time-consuming. In this work, we use reinforcement learning (RL) to find a dynamic policy that selects lab test panels sequentially based on previous observations, ensuring accurate testing at a low cost. Clinical diagnostic data are often highly imbalanced; therefore, we aim to maximize the $F_1$ score instead of the error rate. However, optimizing the non-concave $F_1$ score is not a classic RL problem, thus invalidates standard RL methods. To remedy this issue, we develop a reward shaping approach, leveraging properties of the $F_1$ score and duality of policy optimization, to provably find the set of all Pareto-optimal policies for budget-constrained $F_1$ score maximization. To handle the combinatorially complex state space, we propose a Semi-Model-based Deep Diagnosis Policy Optimization (SM-DDPO) framework that is compatible with end-to-end training and online learning. SM-DDPO is tested on diverse clinical tasks: ferritin abnormality detection, sepsis mortality prediction, and acute kidney injury diagnosis. Experiments with real-world data validate that SM-DDPO trains efficiently and identifies all Pareto-front solutions. Across all tasks, SM-DDPO is able to achieve state-of-the-art diagnosis accuracy (in some cases higher than conventional methods) with up to $85\%$ reduction in testing cost. The code is available at [https://github.com/Zheng321/Deep-Reinforcement-Learning-for-Cost-Effective-Medical-Diagnosis].
A Comparative Study of Pretrained Language Models for Long Clinical Text
Jan 27, 2023



Abstract:Objective: Clinical knowledge enriched transformer models (e.g., ClinicalBERT) have state-of-the-art results on clinical NLP (natural language processing) tasks. One of the core limitations of these transformer models is the substantial memory consumption due to their full self-attention mechanism, which leads to the performance degradation in long clinical texts. To overcome this, we propose to leverage long-sequence transformer models (e.g., Longformer and BigBird), which extend the maximum input sequence length from 512 to 4096, to enhance the ability to model long-term dependencies in long clinical texts. Materials and Methods: Inspired by the success of long sequence transformer models and the fact that clinical notes are mostly long, we introduce two domain enriched language models, Clinical-Longformer and Clinical-BigBird, which are pre-trained on a large-scale clinical corpus. We evaluate both language models using 10 baseline tasks including named entity recognition, question answering, natural language inference, and document classification tasks. Results: The results demonstrate that Clinical-Longformer and Clinical-BigBird consistently and significantly outperform ClinicalBERT and other short-sequence transformers in all 10 downstream tasks and achieve new state-of-the-art results. Discussion: Our pre-trained language models provide the bedrock for clinical NLP using long texts. We have made our source code available at https://github.com/luoyuanlab/Clinical-Longformer, and the pre-trained models available for public download at: https://huggingface.co/yikuan8/Clinical-Longformer. Conclusion: This study demonstrates that clinical knowledge enriched long-sequence transformers are able to learn long-term dependencies in long clinical text. Our methods can also inspire the development of other domain-enriched long-sequence transformers.
Clinical outcome prediction under hypothetical interventions -- a representation learning framework for counterfactual reasoning
May 15, 2022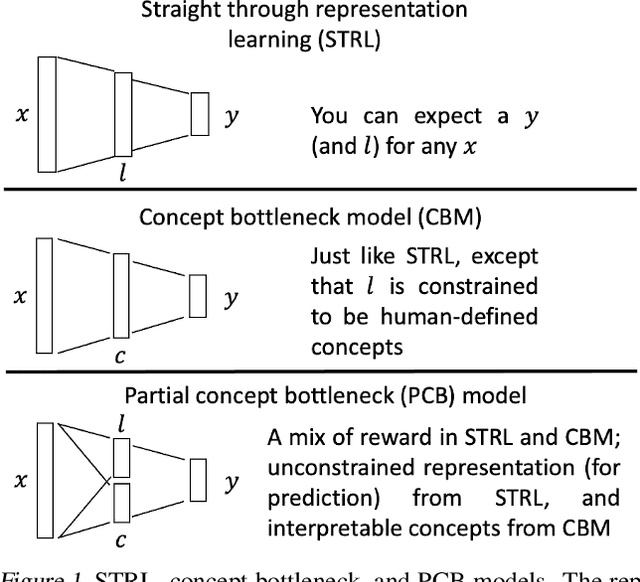
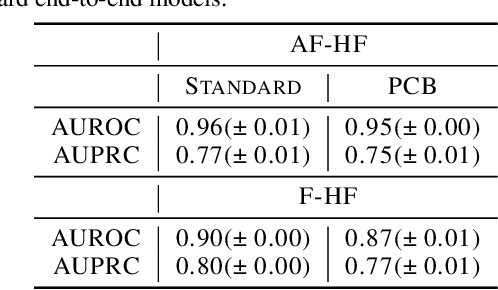
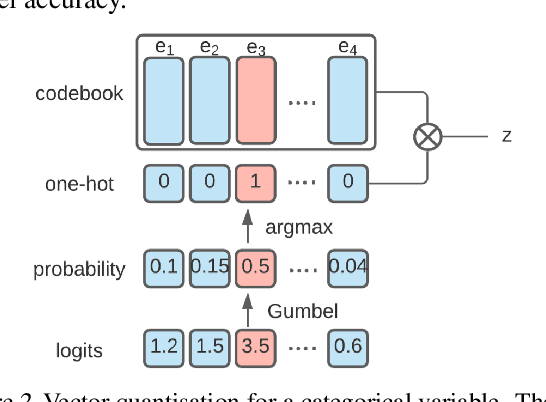
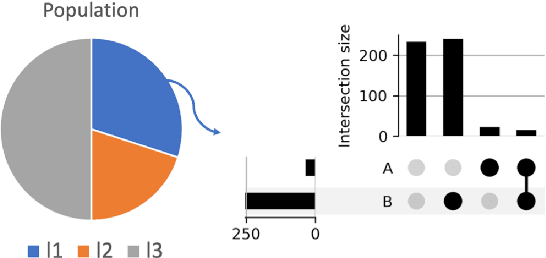
Abstract:Most machine learning (ML) models are developed for prediction only; offering no option for causal interpretation of their predictions or parameters/properties. This can hamper the health systems' ability to employ ML models in clinical decision-making processes, where the need and desire for predicting outcomes under hypothetical investigations (i.e., counterfactual reasoning/explanation) is high. In this research, we introduce a new representation learning framework (i.e., partial concept bottleneck), which considers the provision of counterfactual explanations as an embedded property of the risk model. Despite architectural changes necessary for jointly optimising for prediction accuracy and counterfactual reasoning, the accuracy of our approach is comparable to prediction-only models. Our results suggest that our proposed framework has the potential to help researchers and clinicians improve personalised care (e.g., by investigating the hypothetical differential effects of interventions)
Multimodal Machine Learning in Precision Health
Apr 10, 2022



Abstract:As machine learning and artificial intelligence are more frequently being leveraged to tackle problems in the health sector, there has been increased interest in utilizing them in clinical decision-support. This has historically been the case in single modal data such as electronic health record data. Attempts to improve prediction and resemble the multimodal nature of clinical expert decision-making this has been met in the computational field of machine learning by a fusion of disparate data. This review was conducted to summarize this field and identify topics ripe for future research. We conducted this review in accordance with the PRISMA (Preferred Reporting Items for Systematic reviews and Meta-Analyses) extension for Scoping Reviews to characterize multi-modal data fusion in health. We used a combination of content analysis and literature searches to establish search strings and databases of PubMed, Google Scholar, and IEEEXplore from 2011 to 2021. A final set of 125 articles were included in the analysis. The most common health areas utilizing multi-modal methods were neurology and oncology. However, there exist a wide breadth of current applications. The most common form of information fusion was early fusion. Notably, there was an improvement in predictive performance performing heterogeneous data fusion. Lacking from the papers were clear clinical deployment strategies and pursuit of FDA-approved tools. These findings provide a map of the current literature on multimodal data fusion as applied to health diagnosis/prognosis problems. Multi-modal machine learning, while more robust in its estimations over unimodal methods, has drawbacks in its scalability and the time-consuming nature of information concatenation.
Clinical-Longformer and Clinical-BigBird: Transformers for long clinical sequences
Feb 12, 2022



Abstract:Transformers-based models, such as BERT, have dramatically improved the performance for various natural language processing tasks. The clinical knowledge enriched model, namely ClinicalBERT, also achieved state-of-the-art results when performed on clinical named entity recognition and natural language inference tasks. One of the core limitations of these transformers is the substantial memory consumption due to their full self-attention mechanism. To overcome this, long sequence transformer models, e.g. Longformer and BigBird, were proposed with the idea of sparse attention mechanism to reduce the memory usage from quadratic to the sequence length to a linear scale. These models extended the maximum input sequence length from 512 to 4096, which enhanced the ability of modeling long-term dependency and consequently achieved optimal results in a variety of tasks. Inspired by the success of these long sequence transformer models, we introduce two domain enriched language models, namely Clinical-Longformer and Clinical-BigBird, which are pre-trained from large-scale clinical corpora. We evaluate both pre-trained models using 10 baseline tasks including named entity recognition, question answering, and document classification tasks. The results demonstrate that Clinical-Longformer and Clinical-BigBird consistently and significantly outperform ClinicalBERT as well as other short-sequence transformers in all downstream tasks. We have made our source code available at [https://github.com/luoyuanlab/Clinical-Longformer] the pre-trained models available for public download at: [https://huggingface.co/yikuan8/Clinical-Longformer].
Targeted-BEHRT: Deep learning for observational causal inference on longitudinal electronic health records
Feb 07, 2022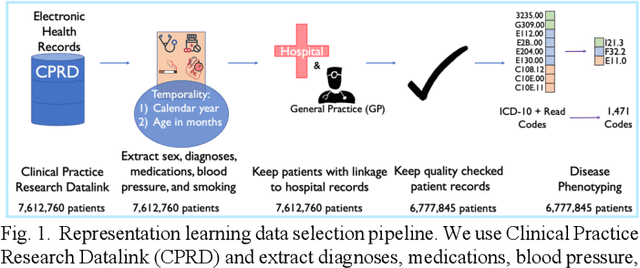
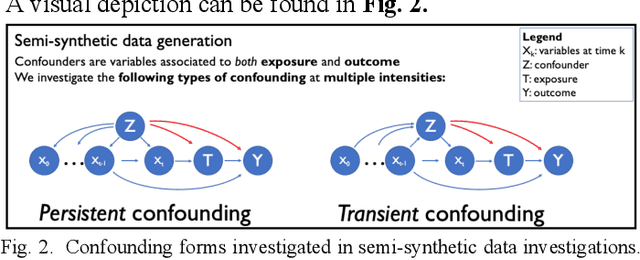
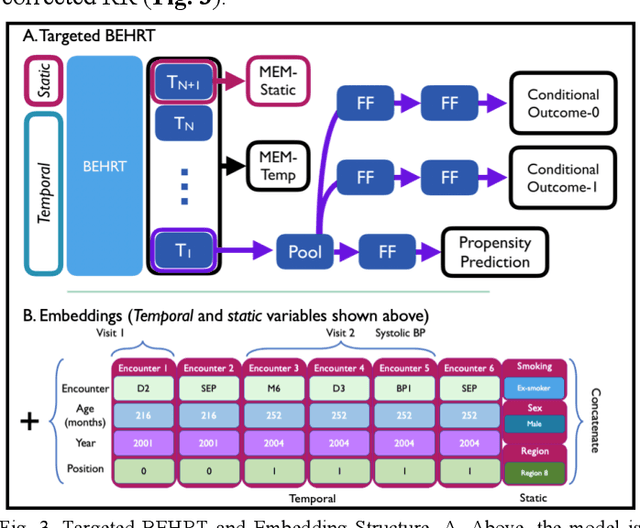
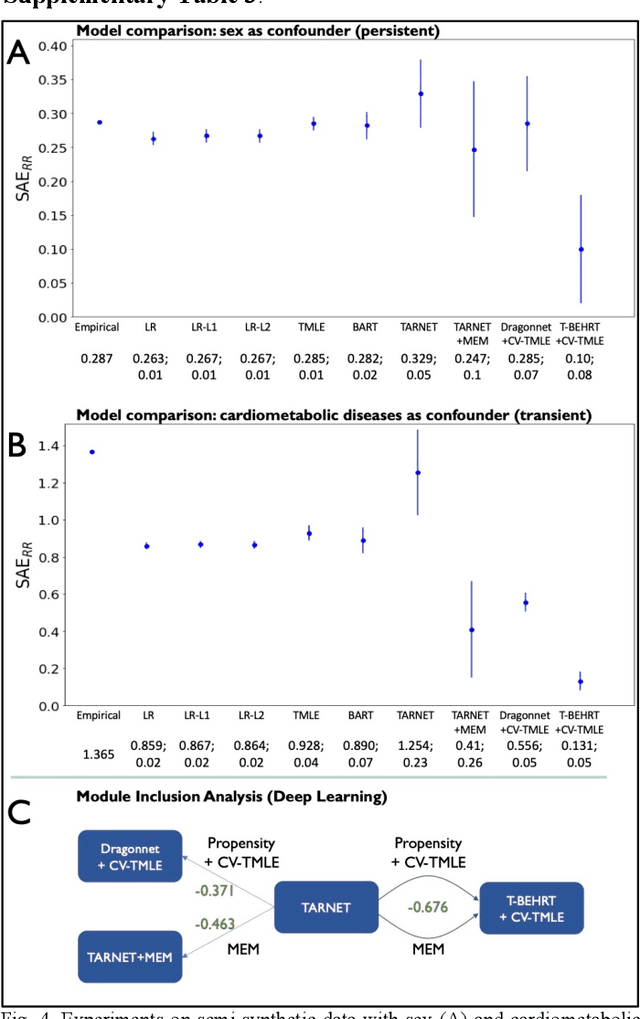
Abstract:Observational causal inference is useful for decision making in medicine when randomized clinical trials (RCT) are infeasible or non generalizable. However, traditional approaches fail to deliver unconfounded causal conclusions in practice. The rise of "doubly robust" non-parametric tools coupled with the growth of deep learning for capturing rich representations of multimodal data, offers a unique opportunity to develop and test such models for causal inference on comprehensive electronic health records (EHR). In this paper, we investigate causal modelling of an RCT-established null causal association: the effect of antihypertensive use on incident cancer risk. We develop a dataset for our observational study and a Transformer-based model, Targeted BEHRT coupled with doubly robust estimation, we estimate average risk ratio (RR). We compare our model to benchmark statistical and deep learning models for causal inference in multiple experiments on semi-synthetic derivations of our dataset with various types and intensities of confounding. In order to further test the reliability of our approach, we test our model on situations of limited data. We find that our model provides more accurate estimates of RR (least sum absolute error from ground truth) compared to benchmarks for risk ratio estimation on high-dimensional EHR across experiments. Finally, we apply our model to investigate the original case study: antihypertensives' effect on cancer and demonstrate that our model generally captures the validated null association.
Disparities in Social Determinants among Performances of Mortality Prediction with Machine Learning for Sepsis Patients
Dec 15, 2021



Abstract:Background Sepsis is one of the most life-threatening circumstances for critically ill patients in the US, while a standardized criteria for sepsis identification is still under development. Disparities in social determinants of sepsis patients can interfere with the risk prediction performances using machine learning. Methods Disparities in social determinants, including race, gender, marital status, insurance types and languages, among patients identified by six available sepsis criteria were revealed by forest plots. Sixteen machine learning classifiers were trained to predict in-hospital mortality for sepsis patients. The performance of the trained model was tested on the entire randomly conducted test set and each sub-population built based on each of the following social determinants: race, gender, marital status, insurance type, and language. Results We analyzed a total of 11,791 critical care patients from the MIMIC-III database. Within the population identified by each sepsis identification method, significant differences were observed among sub-populations regarding race, marital status, insurance type, and language. On the 5,783 sepsis patients identified by the Sepsis-3 criteria statistically significant performance decreases for mortality prediction were observed when applying the trained machine learning model on Asian and Hispanic patients. With pairwise comparison, we detected performance discrepancies in mortality prediction between Asian and White patients, Asians and patients of other races, as well as English-speaking and Spanish-speaking patients. Conclusions Disparities in proportions of patients identified by various sepsis criteria were detected among the different social determinant groups. To achieve accurate diagnosis, a versatile diagnostic system for sepsis is needed to overcome the social determinant disparities of patients.
Early Prediction of Mortality in Critical Care Setting in Sepsis Patients Using Structured Features and Unstructured Clinical Notes
Nov 09, 2021
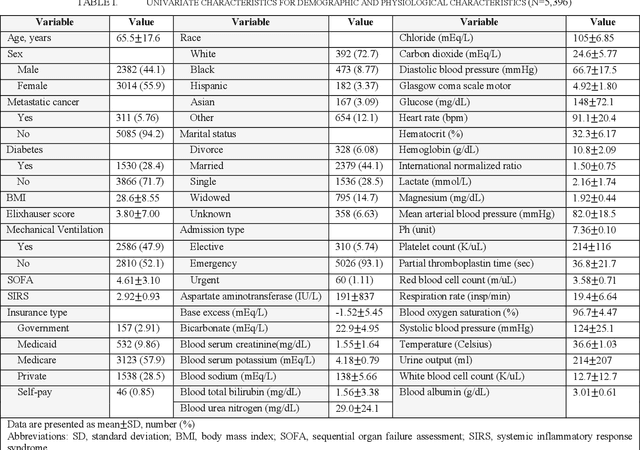
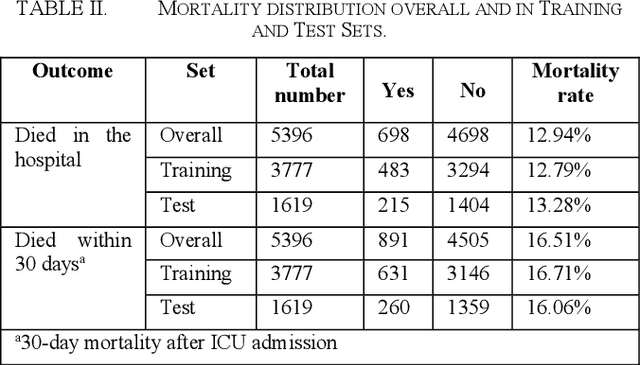
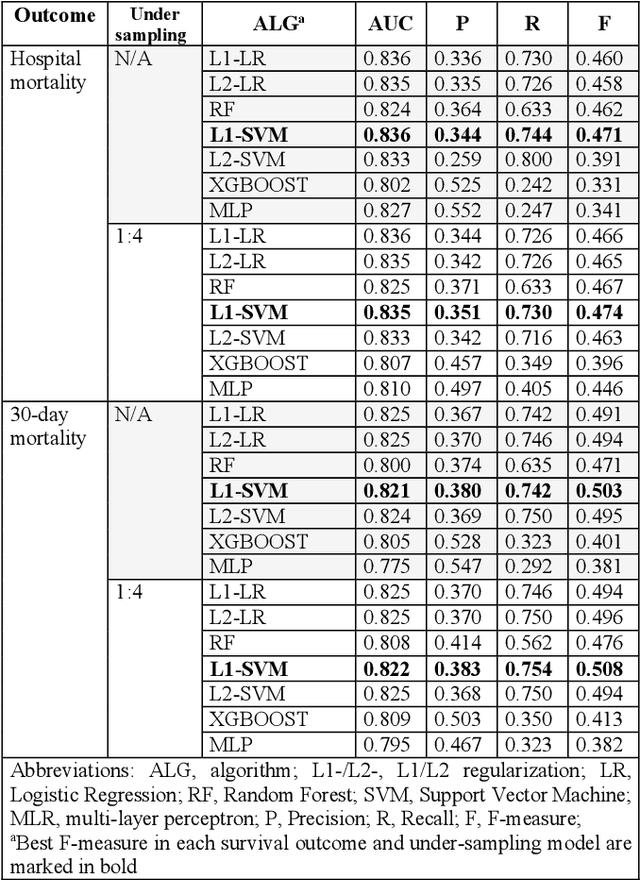
Abstract:Sepsis is an important cause of mortality, especially in intensive care unit (ICU) patients. Developing novel methods to identify early mortality is critical for improving survival outcomes in sepsis patients. Using the MIMIC-III database, we integrated demographic data, physiological measurements and clinical notes. We built and applied several machine learning models to predict the risk of hospital mortality and 30-day mortality in sepsis patients. From the clinical notes, we generated clinically meaningful word representations and embeddings. Supervised learning classifiers and a deep learning architecture were used to construct prediction models. The configurations that utilized both structured and unstructured clinical features yielded competitive F-measure of 0.512. Our results showed that the approaches integrating both structured and unstructured clinical features can be effectively applied to assist clinicians in identifying the risk of mortality in sepsis patients upon admission to the ICU.
Transfer Learning in Electronic Health Records through Clinical Concept Embedding
Jul 27, 2021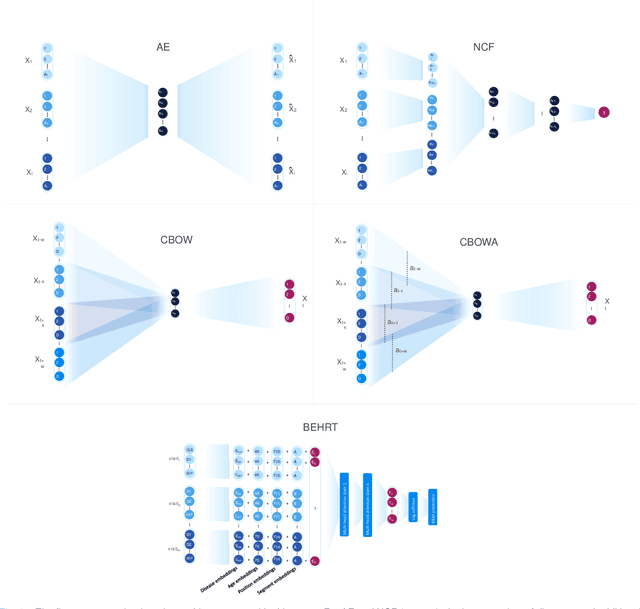
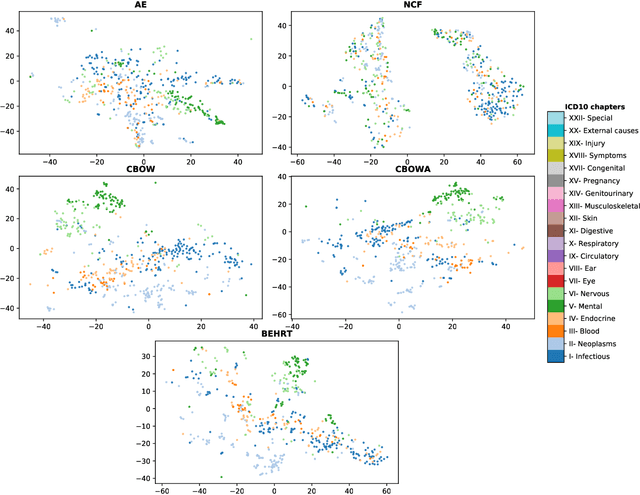
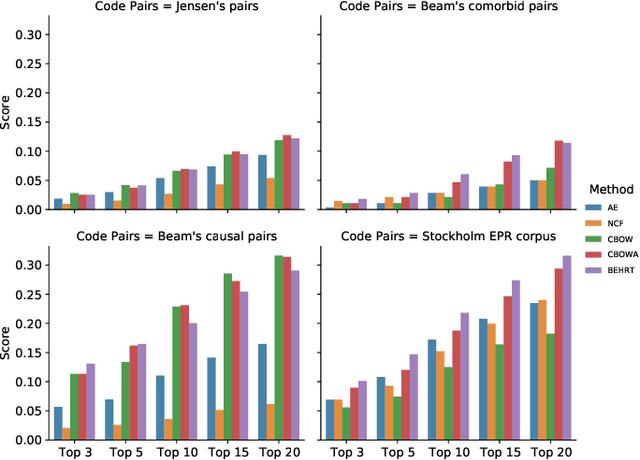
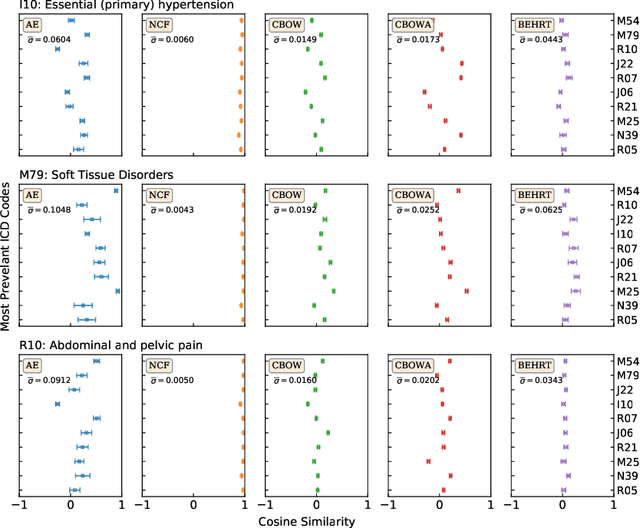
Abstract:Deep learning models have shown tremendous potential in learning representations, which are able to capture some key properties of the data. This makes them great candidates for transfer learning: Exploiting commonalities between different learning tasks to transfer knowledge from one task to another. Electronic health records (EHR) research is one of the domains that has witnessed a growing number of deep learning techniques employed for learning clinically-meaningful representations of medical concepts (such as diseases and medications). Despite this growth, the approaches to benchmark and assess such learned representations (or, embeddings) is under-investigated; this can be a big issue when such embeddings are shared to facilitate transfer learning. In this study, we aim to (1) train some of the most prominent disease embedding techniques on a comprehensive EHR data from 3.1 million patients, (2) employ qualitative and quantitative evaluation techniques to assess these embeddings, and (3) provide pre-trained disease embeddings for transfer learning. This study can be the first comprehensive approach for clinical concept embedding evaluation and can be applied to any embedding techniques and for any EHR concept.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge