Gholamreza Salimi-Khorshidi
A Transformer-based survival model for prediction of all-cause mortality in heart failure patients: a multi-cohort study
Mar 16, 2025Abstract:We developed and validated TRisk, a Transformer-based AI model predicting 36-month mortality in heart failure patients by analysing temporal patient journeys from UK electronic health records (EHR). Our study included 403,534 heart failure patients (ages 40-90) from 1,418 English general practices, with 1,063 practices for model derivation and 355 for external validation. TRisk was compared against the MAGGIC-EHR model across various patient subgroups. With median follow-up of 9 months, TRisk achieved a concordance index of 0.845 (95% confidence interval: [0.841, 0.849]), significantly outperforming MAGGIC-EHR's 0.728 (0.723, 0.733) for predicting 36-month all-cause mortality. TRisk showed more consistent performance across sex, age, and baseline characteristics, suggesting less bias. We successfully adapted TRisk to US hospital data through transfer learning, achieving a C-index of 0.802 (0.789, 0.816) with 21,767 patients. Explainability analyses revealed TRisk captured established risk factors while identifying underappreciated predictors like cancers and hepatic failure that were important across both cohorts. Notably, cancers maintained strong prognostic value even a decade after diagnosis. TRisk demonstrated well-calibrated mortality prediction across both healthcare systems. Our findings highlight the value of tracking longitudinal health profiles and revealed risk factors not included in previous expert-driven models.
Clinical outcome prediction under hypothetical interventions -- a representation learning framework for counterfactual reasoning
May 15, 2022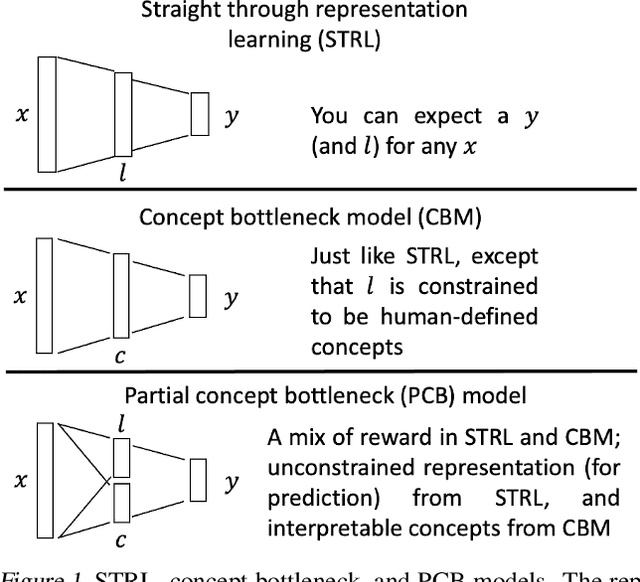
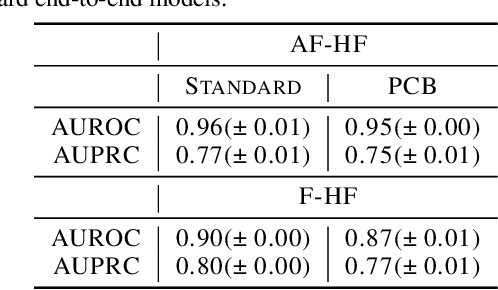
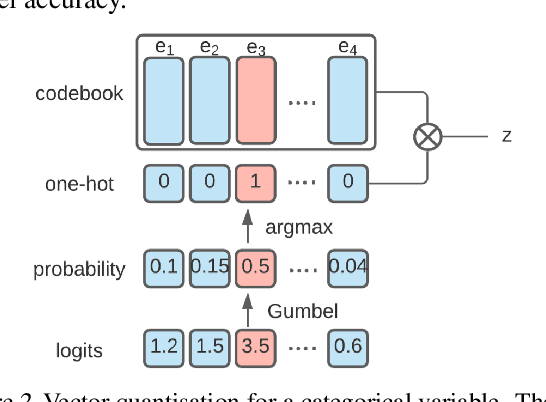
Abstract:Most machine learning (ML) models are developed for prediction only; offering no option for causal interpretation of their predictions or parameters/properties. This can hamper the health systems' ability to employ ML models in clinical decision-making processes, where the need and desire for predicting outcomes under hypothetical investigations (i.e., counterfactual reasoning/explanation) is high. In this research, we introduce a new representation learning framework (i.e., partial concept bottleneck), which considers the provision of counterfactual explanations as an embedded property of the risk model. Despite architectural changes necessary for jointly optimising for prediction accuracy and counterfactual reasoning, the accuracy of our approach is comparable to prediction-only models. Our results suggest that our proposed framework has the potential to help researchers and clinicians improve personalised care (e.g., by investigating the hypothetical differential effects of interventions)
Targeted-BEHRT: Deep learning for observational causal inference on longitudinal electronic health records
Feb 07, 2022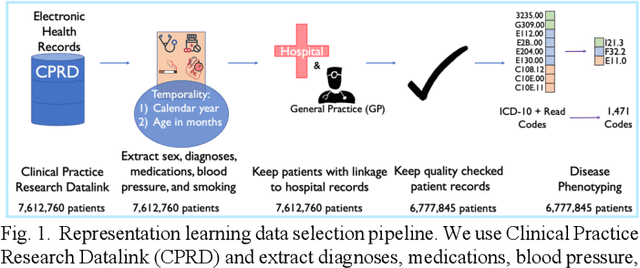
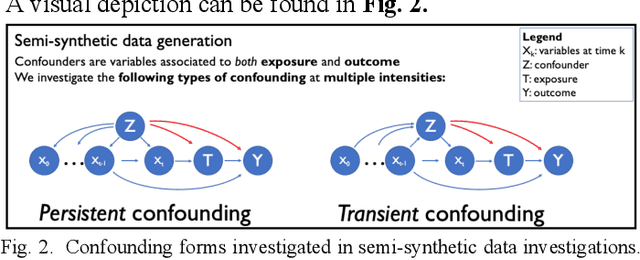
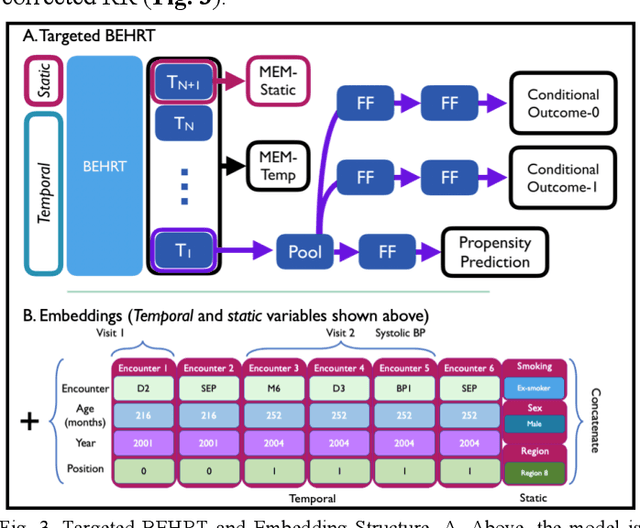
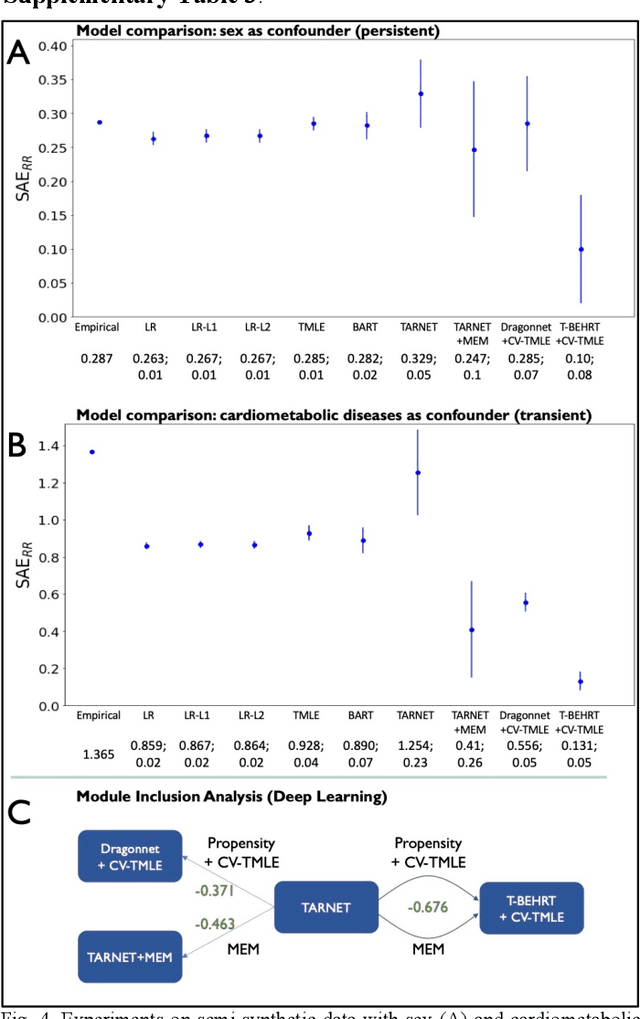
Abstract:Observational causal inference is useful for decision making in medicine when randomized clinical trials (RCT) are infeasible or non generalizable. However, traditional approaches fail to deliver unconfounded causal conclusions in practice. The rise of "doubly robust" non-parametric tools coupled with the growth of deep learning for capturing rich representations of multimodal data, offers a unique opportunity to develop and test such models for causal inference on comprehensive electronic health records (EHR). In this paper, we investigate causal modelling of an RCT-established null causal association: the effect of antihypertensive use on incident cancer risk. We develop a dataset for our observational study and a Transformer-based model, Targeted BEHRT coupled with doubly robust estimation, we estimate average risk ratio (RR). We compare our model to benchmark statistical and deep learning models for causal inference in multiple experiments on semi-synthetic derivations of our dataset with various types and intensities of confounding. In order to further test the reliability of our approach, we test our model on situations of limited data. We find that our model provides more accurate estimates of RR (least sum absolute error from ground truth) compared to benchmarks for risk ratio estimation on high-dimensional EHR across experiments. Finally, we apply our model to investigate the original case study: antihypertensives' effect on cancer and demonstrate that our model generally captures the validated null association.
Transfer Learning in Electronic Health Records through Clinical Concept Embedding
Jul 27, 2021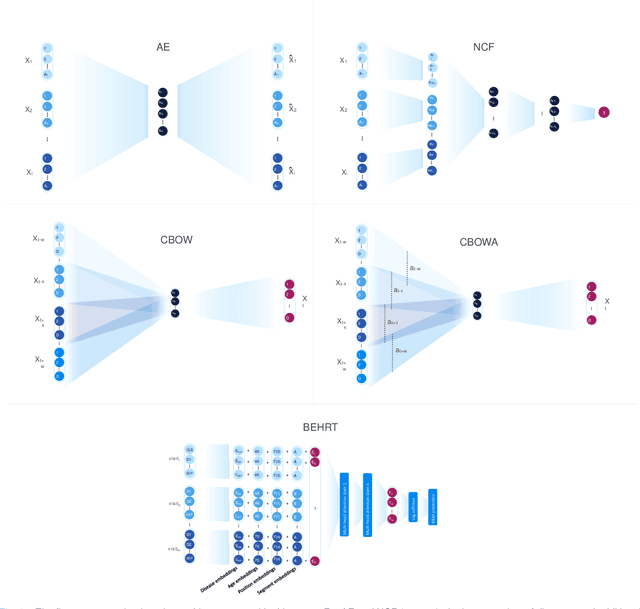
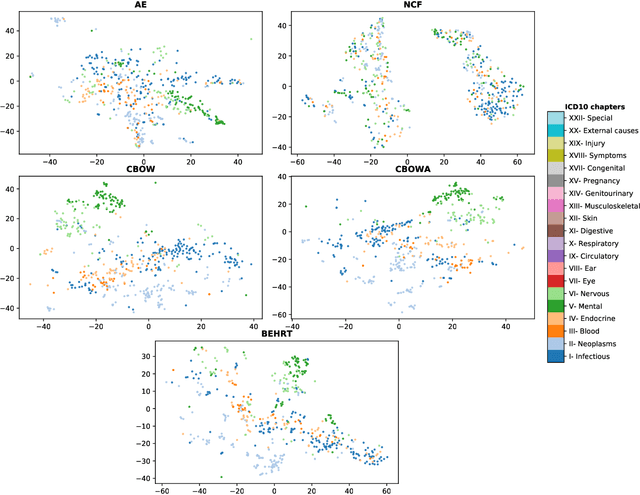
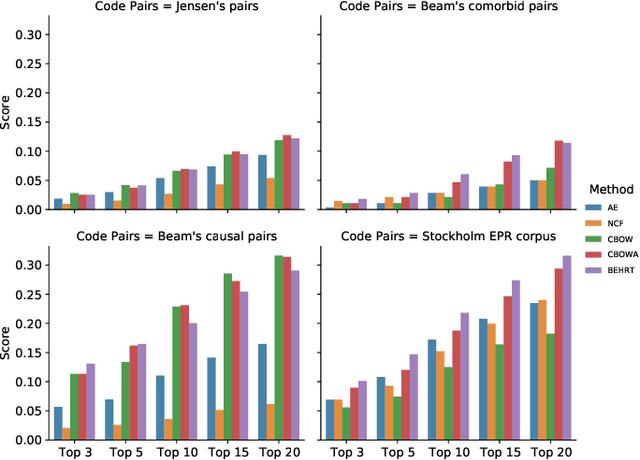
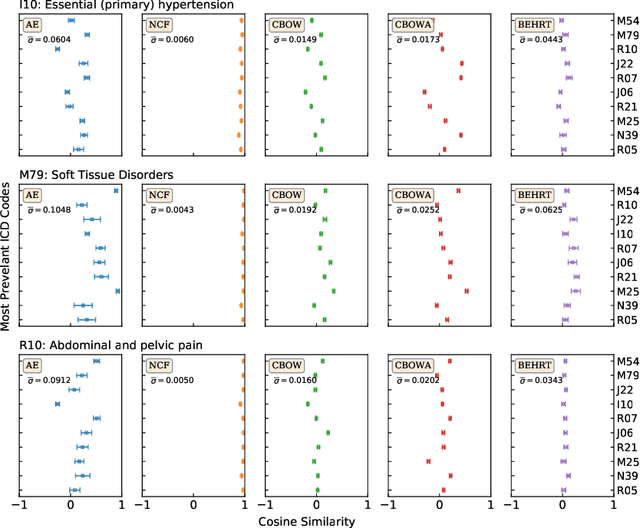
Abstract:Deep learning models have shown tremendous potential in learning representations, which are able to capture some key properties of the data. This makes them great candidates for transfer learning: Exploiting commonalities between different learning tasks to transfer knowledge from one task to another. Electronic health records (EHR) research is one of the domains that has witnessed a growing number of deep learning techniques employed for learning clinically-meaningful representations of medical concepts (such as diseases and medications). Despite this growth, the approaches to benchmark and assess such learned representations (or, embeddings) is under-investigated; this can be a big issue when such embeddings are shared to facilitate transfer learning. In this study, we aim to (1) train some of the most prominent disease embedding techniques on a comprehensive EHR data from 3.1 million patients, (2) employ qualitative and quantitative evaluation techniques to assess these embeddings, and (3) provide pre-trained disease embeddings for transfer learning. This study can be the first comprehensive approach for clinical concept embedding evaluation and can be applied to any embedding techniques and for any EHR concept.
Hi-BEHRT: Hierarchical Transformer-based model for accurate prediction of clinical events using multimodal longitudinal electronic health records
Jun 21, 2021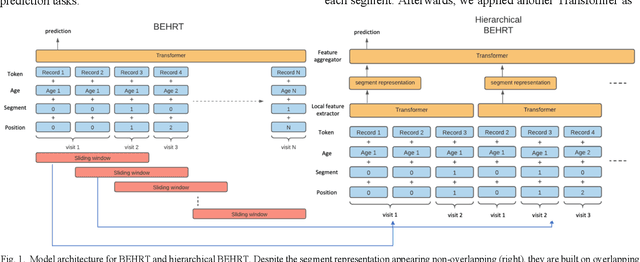
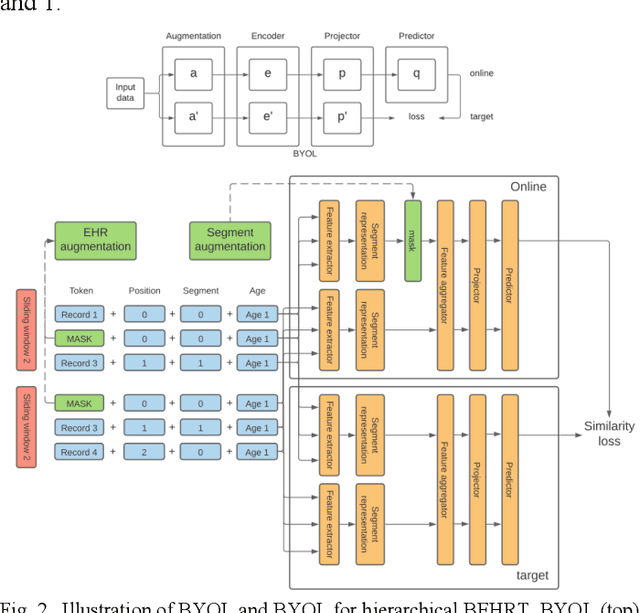
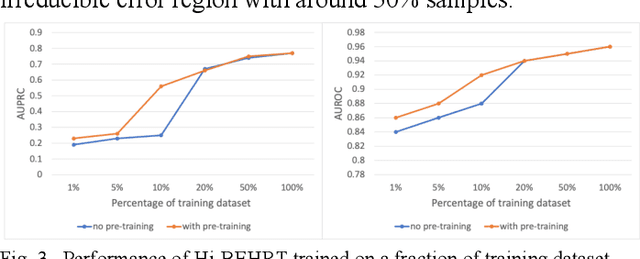
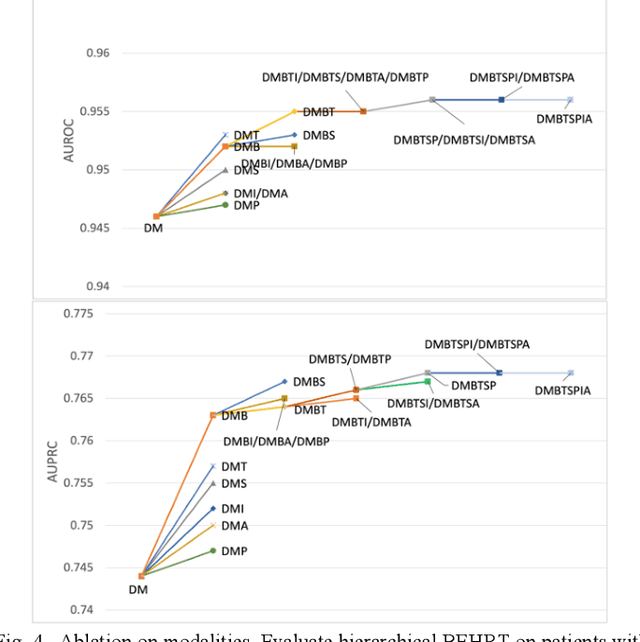
Abstract:Electronic health records represent a holistic overview of patients' trajectories. Their increasing availability has fueled new hopes to leverage them and develop accurate risk prediction models for a wide range of diseases. Given the complex interrelationships of medical records and patient outcomes, deep learning models have shown clear merits in achieving this goal. However, a key limitation of these models remains their capacity in processing long sequences. Capturing the whole history of medical encounters is expected to lead to more accurate predictions, but the inclusion of records collected for decades and from multiple resources can inevitably exceed the receptive field of the existing deep learning architectures. This can result in missing crucial, long-term dependencies. To address this gap, we present Hi-BEHRT, a hierarchical Transformer-based model that can significantly expand the receptive field of Transformers and extract associations from much longer sequences. Using a multimodal large-scale linked longitudinal electronic health records, the Hi-BEHRT exceeds the state-of-the-art BEHRT 1% to 5% for area under the receiver operating characteristic (AUROC) curve and 3% to 6% for area under the precision recall (AUPRC) curve on average, and 3% to 6% (AUROC) and 3% to 11% (AUPRC) for patients with long medical history for 5-year heart failure, diabetes, chronic kidney disease, and stroke risk prediction. Additionally, because pretraining for hierarchical Transformer is not well-established, we provide an effective end-to-end contrastive pre-training strategy for Hi-BEHRT using EHR, improving its transferability on predicting clinical events with relatively small training dataset.
Risk factor identification for incident heart failure using neural network distillation and variable selection
Mar 01, 2021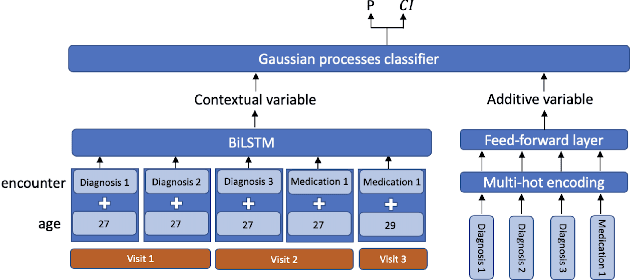
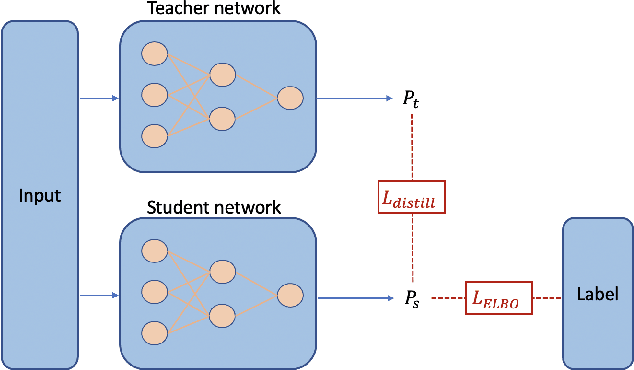
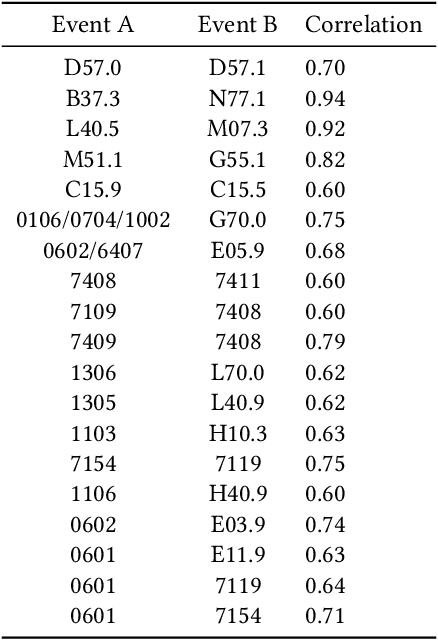
Abstract:Recent evidence shows that deep learning models trained on electronic health records from millions of patients can deliver substantially more accurate predictions of risk compared to their statistical counterparts. While this provides an important opportunity for improving clinical decision-making, the lack of interpretability is a major barrier to the incorporation of these black-box models in routine care, limiting their trustworthiness and preventing further hypothesis-testing investigations. In this study, we propose two methods, namely, model distillation and variable selection, to untangle hidden patterns learned by an established deep learning model (BEHRT) for risk association identification. Due to the clinical importance and diversity of heart failure as a phenotype, it was used to showcase the merits of the proposed methods. A cohort with 788,880 (8.3% incident heart failure) patients was considered for the study. Model distillation identified 598 and 379 diseases that were associated and dissociated with heart failure at the population level, respectively. While the associations were broadly consistent with prior knowledge, our method also highlighted several less appreciated links that are worth further investigation. In addition to these important population-level insights, we developed an approach to individual-level interpretation to take account of varying manifestation of heart failure in clinical practice. This was achieved through variable selection by detecting a minimal set of encounters that can maximally preserve the accuracy of prediction for individuals. Our proposed work provides a discovery-enabling tool to identify risk factors in both population and individual levels from a data-driven perspective. This helps to generate new hypotheses and guides further investigations on causal links.
An explainable Transformer-based deep learning model for the prediction of incident heart failure
Jan 27, 2021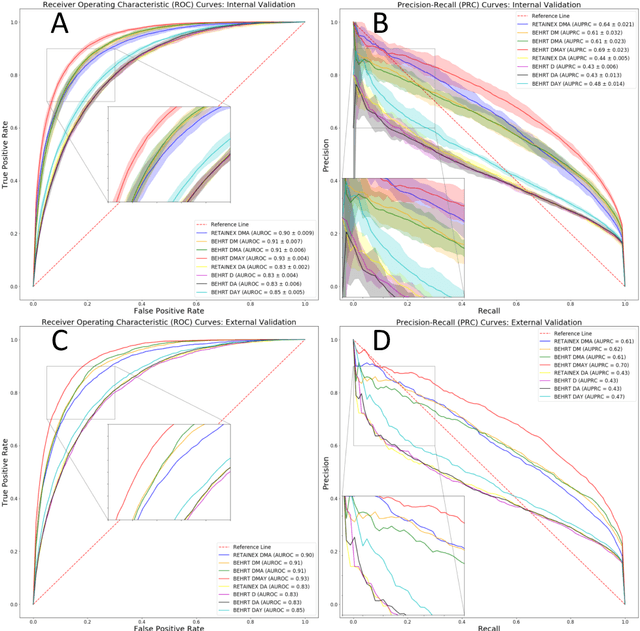
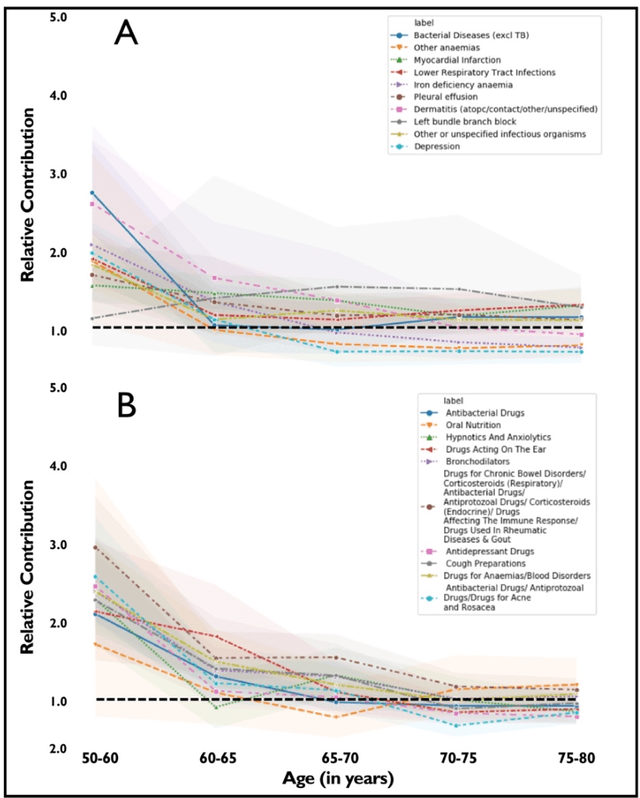
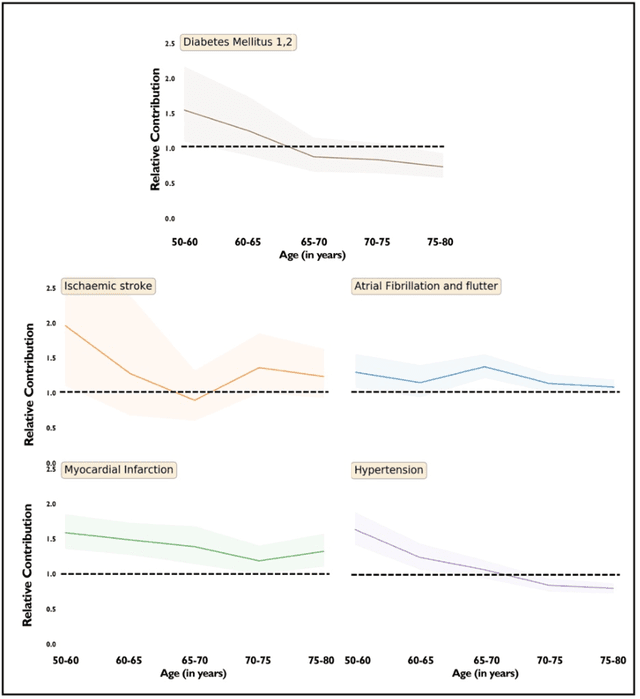
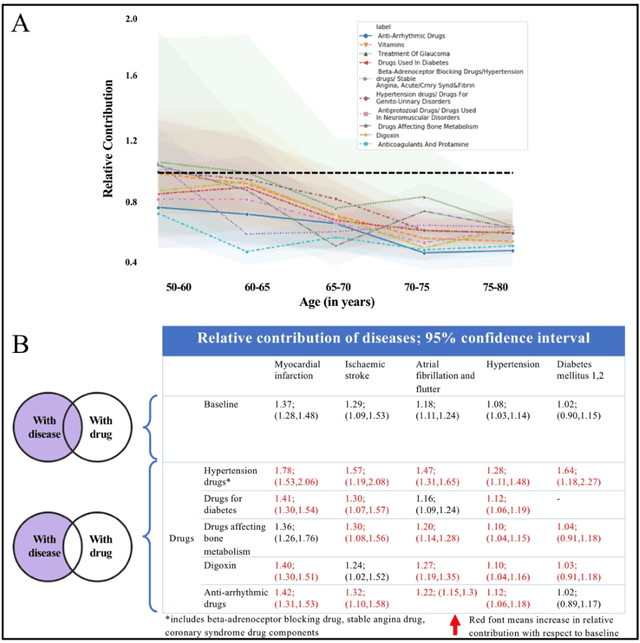
Abstract:Predicting the incidence of complex chronic conditions such as heart failure is challenging. Deep learning models applied to rich electronic health records may improve prediction but remain unexplainable hampering their wider use in medical practice. We developed a novel Transformer deep-learning model for more accurate and yet explainable prediction of incident heart failure involving 100,071 patients from longitudinal linked electronic health records across the UK. On internal 5-fold cross validation and held-out external validation, our model achieved 0.93 and 0.93 area under the receiver operator curve and 0.69 and 0.70 area under the precision-recall curve, respectively and outperformed existing deep learning models. Predictor groups included all community and hospital diagnoses and medications contextualised within the age and calendar year for each patient's clinical encounter. The importance of contextualised medical information was revealed in a number of sensitivity analyses, and our perturbation method provided a way of identifying factors contributing to risk. Many of the identified risk factors were consistent with existing knowledge from clinical and epidemiological research but several new associations were revealed which had not been considered in expert-driven risk prediction models.
Deep Bayesian Gaussian Processes for Uncertainty Estimation in Electronic Health Records
Mar 23, 2020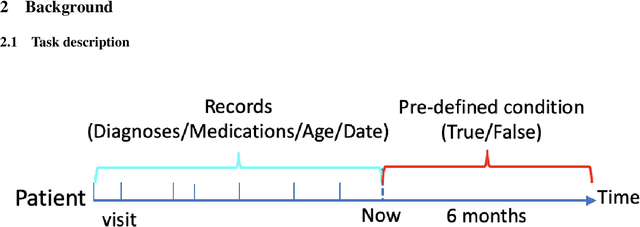
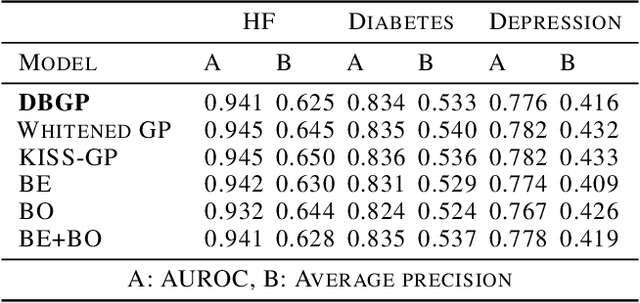
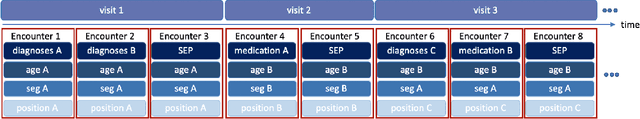
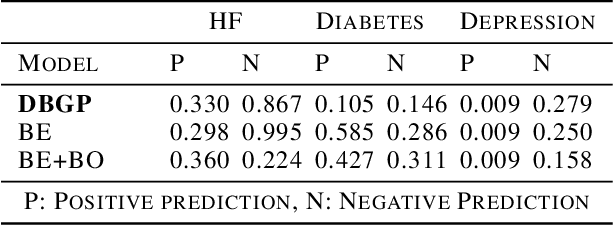
Abstract:One major impediment to the wider use of deep learning for clinical decision making is the difficulty of assigning a level of confidence to model predictions. Currently, deep Bayesian neural networks and sparse Gaussian processes are the main two scalable uncertainty estimation methods. However, deep Bayesian neural network suffers from lack of expressiveness, and more expressive models such as deep kernel learning, which is an extension of sparse Gaussian process, captures only the uncertainty from the higher level latent space. Therefore, the deep learning model under it lacks interpretability and ignores uncertainty from the raw data. In this paper, we merge features of the deep Bayesian learning framework with deep kernel learning to leverage the strengths of both methods for more comprehensive uncertainty estimation. Through a series of experiments on predicting the first incidence of heart failure, diabetes and depression applied to large-scale electronic medical records, we demonstrate that our method is better at capturing uncertainty than both Gaussian processes and deep Bayesian neural networks in terms of indicating data insufficiency and distinguishing true positive and false positive predictions, with a comparable generalisation performance. Furthermore, by assessing the accuracy and area under the receiver operating characteristic curve over the predictive probability, we show that our method is less susceptible to making overconfident predictions, especially for the minority class in imbalanced datasets. Finally, we demonstrate how uncertainty information derived by the model can inform risk factor analysis towards model interpretability.
Topology Distance: A Topology-Based Approach For Evaluating Generative Adversarial Networks
Feb 27, 2020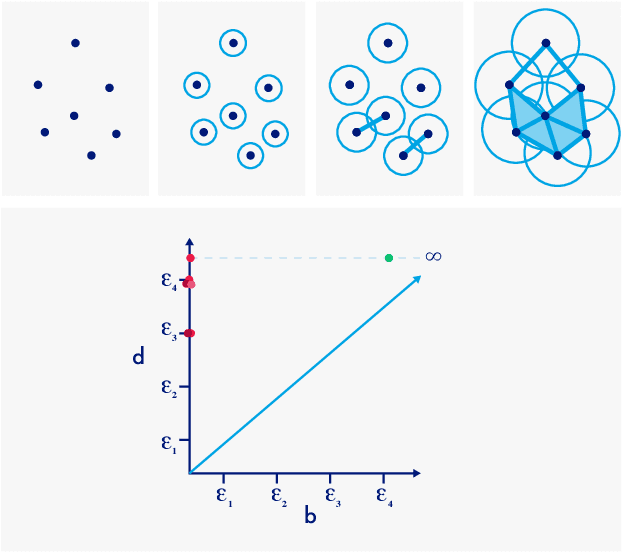
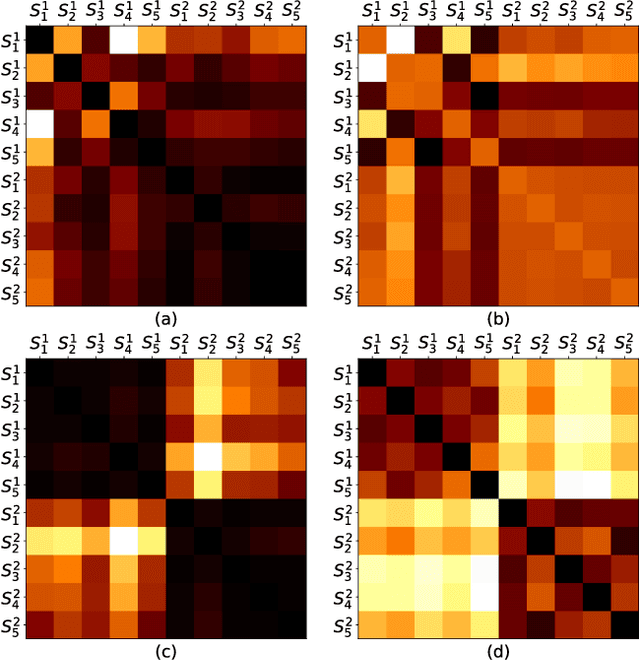
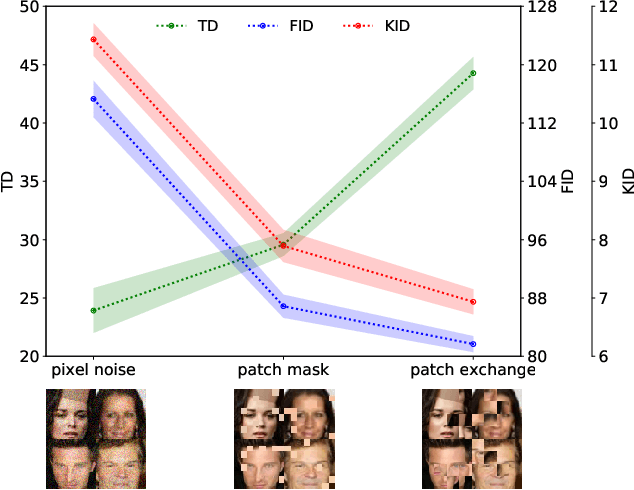

Abstract:Automatic evaluation of the goodness of Generative Adversarial Networks (GANs) has been a challenge for the field of machine learning. In this work, we propose a distance complementary to existing measures: Topology Distance (TD), the main idea behind which is to compare the geometric and topological features of the latent manifold of real data with those of generated data. More specifically, we build Vietoris-Rips complex on image features, and define TD based on the differences in persistent-homology groups of the two manifolds. We compare TD with the most commonly used and relevant measures in the field, including Inception Score (IS), Frechet Inception Distance (FID), Kernel Inception Distance (KID) and Geometry Score (GS), in a range of experiments on various datasets. We demonstrate the unique advantage and superiority of our proposed approach over the aforementioned metrics. A combination of our empirical results and the theoretical argument we propose in favour of TD, strongly supports the claim that TD is a powerful candidate metric that researchers can employ when aiming to automatically evaluate the goodness of GANs' learning.
$¶$ILCRO: Making Importance Landscapes Flat Again
Feb 06, 2020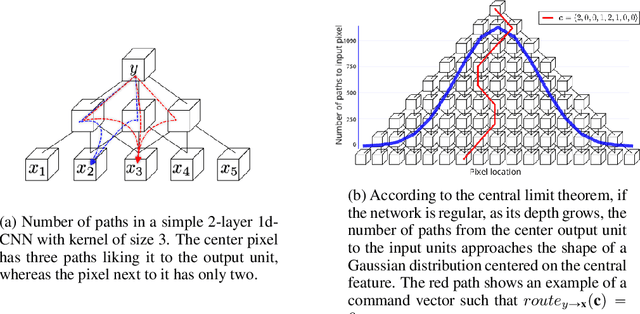
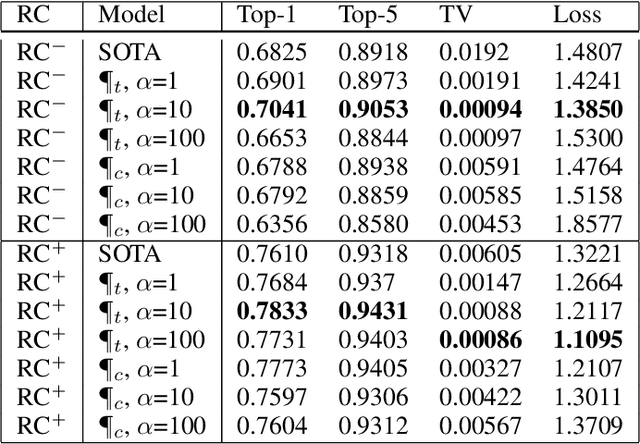
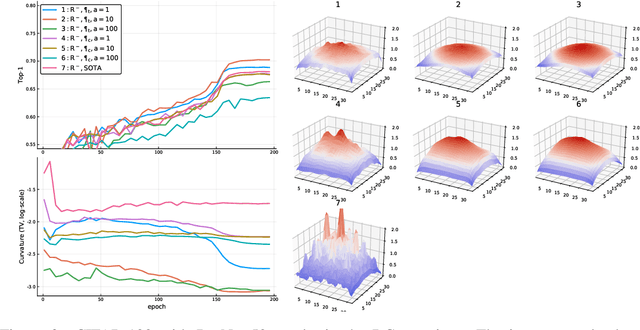
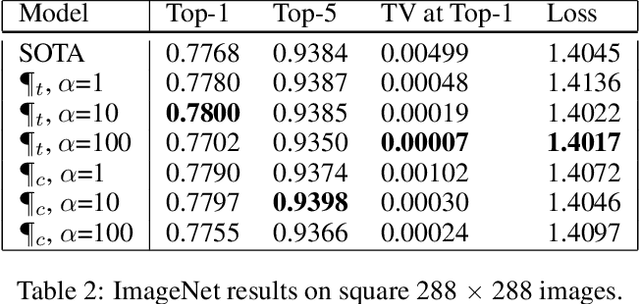
Abstract:Convolutional neural networks have had a great success in numerous tasks, including image classification, object detection, sequence modelling, and many more. It is generally assumed that such neural networks are translation invariant, meaning that they can detect a given feature independent of its location in the input image. While this is true for simple cases, where networks are composed of a restricted number of layer classes and where images are fairly simple, complex images with common state-of-the-art networks do not usually enjoy this property as one might hope. This paper shows that most of the existing convolutional architectures define, at initialisation, a specific feature importance landscape that conditions their capacity to attend to different locations of the images later during training or even at test time. We demonstrate how this phenomenon occurs under specific conditions and how it can be adjusted under some assumptions. We derive the P-objective, or PILCRO for Pixel-wise Importance Landscape Curvature Regularised Objective, a simple regularisation technique that favours weight configurations that produce smooth, low-curvature importance landscapes that are conditioned on the data and not on the chosen architecture. Through extensive experiments, we further show that P-regularised versions of popular computer vision networks have a flat importance landscape, train faster, result in a better accuracy and are more robust to noise at test time, when compared to their original counterparts in common computer-vision classification settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge