Yajie Zhu
Transfer Learning in Electronic Health Records through Clinical Concept Embedding
Jul 27, 2021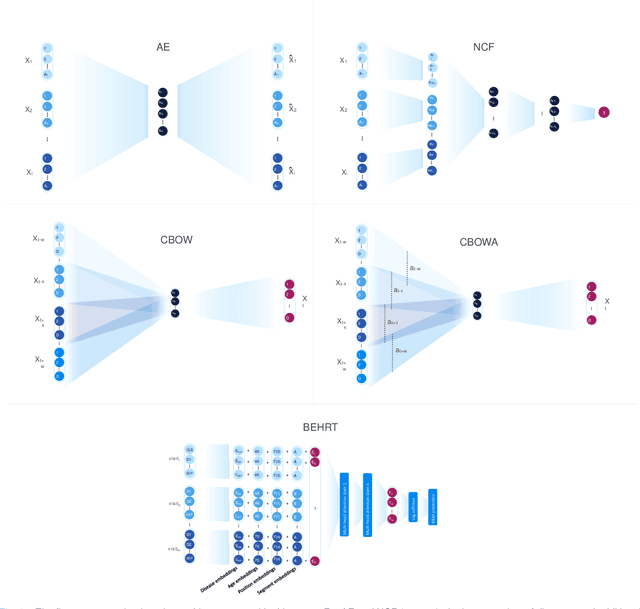
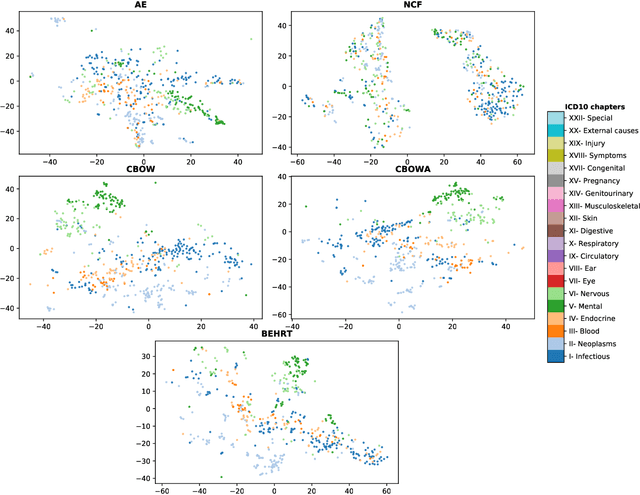
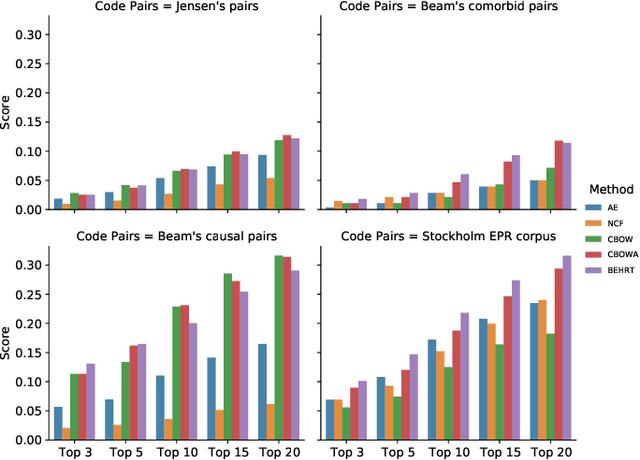
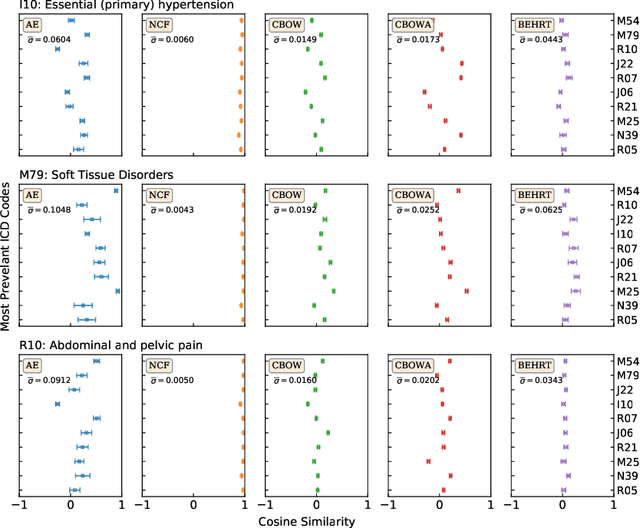
Abstract:Deep learning models have shown tremendous potential in learning representations, which are able to capture some key properties of the data. This makes them great candidates for transfer learning: Exploiting commonalities between different learning tasks to transfer knowledge from one task to another. Electronic health records (EHR) research is one of the domains that has witnessed a growing number of deep learning techniques employed for learning clinically-meaningful representations of medical concepts (such as diseases and medications). Despite this growth, the approaches to benchmark and assess such learned representations (or, embeddings) is under-investigated; this can be a big issue when such embeddings are shared to facilitate transfer learning. In this study, we aim to (1) train some of the most prominent disease embedding techniques on a comprehensive EHR data from 3.1 million patients, (2) employ qualitative and quantitative evaluation techniques to assess these embeddings, and (3) provide pre-trained disease embeddings for transfer learning. This study can be the first comprehensive approach for clinical concept embedding evaluation and can be applied to any embedding techniques and for any EHR concept.
Deep Bayesian Gaussian Processes for Uncertainty Estimation in Electronic Health Records
Mar 23, 2020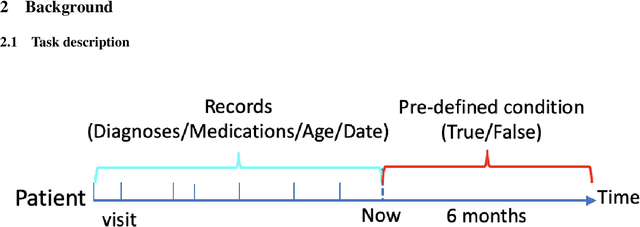
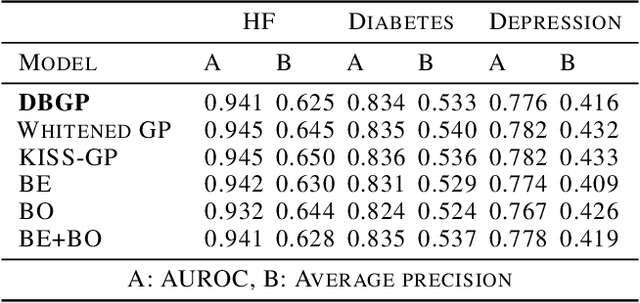
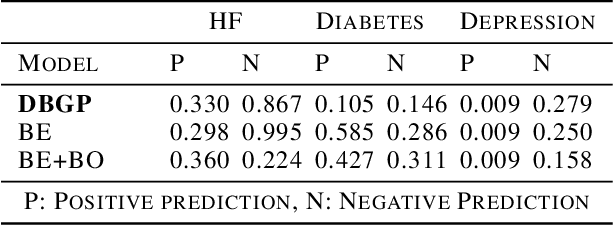
Abstract:One major impediment to the wider use of deep learning for clinical decision making is the difficulty of assigning a level of confidence to model predictions. Currently, deep Bayesian neural networks and sparse Gaussian processes are the main two scalable uncertainty estimation methods. However, deep Bayesian neural network suffers from lack of expressiveness, and more expressive models such as deep kernel learning, which is an extension of sparse Gaussian process, captures only the uncertainty from the higher level latent space. Therefore, the deep learning model under it lacks interpretability and ignores uncertainty from the raw data. In this paper, we merge features of the deep Bayesian learning framework with deep kernel learning to leverage the strengths of both methods for more comprehensive uncertainty estimation. Through a series of experiments on predicting the first incidence of heart failure, diabetes and depression applied to large-scale electronic medical records, we demonstrate that our method is better at capturing uncertainty than both Gaussian processes and deep Bayesian neural networks in terms of indicating data insufficiency and distinguishing true positive and false positive predictions, with a comparable generalisation performance. Furthermore, by assessing the accuracy and area under the receiver operating characteristic curve over the predictive probability, we show that our method is less susceptible to making overconfident predictions, especially for the minority class in imbalanced datasets. Finally, we demonstrate how uncertainty information derived by the model can inform risk factor analysis towards model interpretability.
BEHRT: Transformer for Electronic Health Records
Jul 22, 2019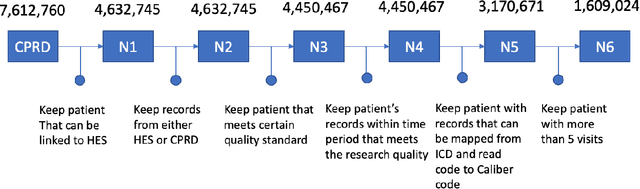

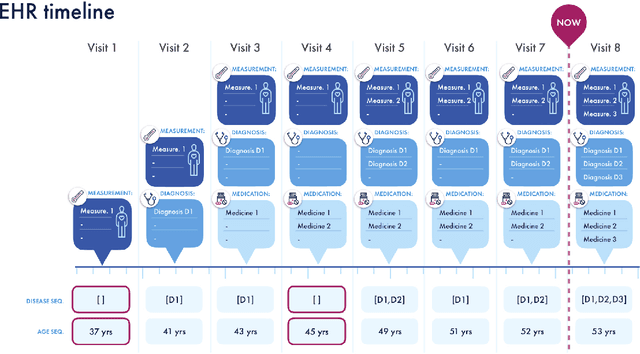
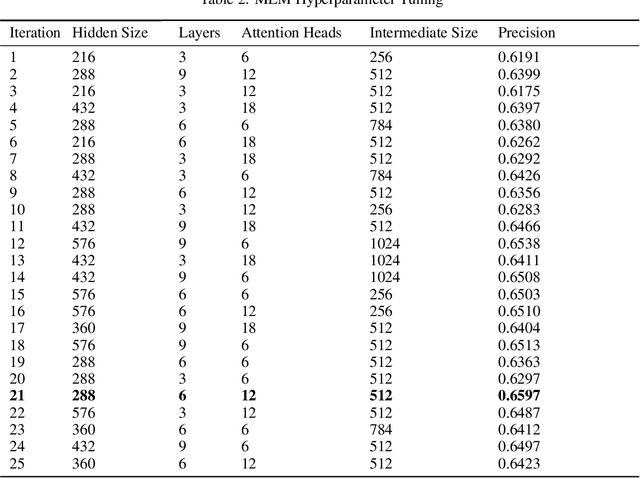
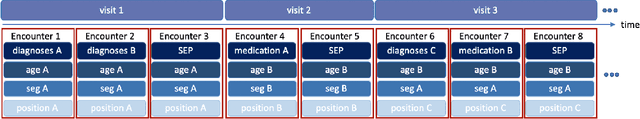
Abstract:Today, despite decades of developments in medicine and the growing interest in precision healthcare, vast majority of diagnoses happen once patients begin to show noticeable signs of illness. Early indication and detection of diseases, however, can provide patients and carers with the chance of early intervention, better disease management, and efficient allocation of healthcare resources. The latest developments in machine learning (more specifically, deep learning) provides a great opportunity to address this unmet need. In this study, we introduce BEHRT: A deep neural sequence transduction model for EHR (electronic health records), capable of multitask prediction and disease trajectory mapping. When trained and evaluated on the data from nearly 1.6 million individuals, BEHRT shows a striking absolute improvement of 8.0-10.8%, in terms of Average Precision Score, compared to the existing state-of-the-art deep EHR models (in terms of average precision, when predicting for the onset of 301 conditions). In addition to its superior prediction power, BEHRT provides a personalised view of disease trajectories through its attention mechanism; its flexible architecture enables it to incorporate multiple heterogeneous concepts (e.g., diagnosis, medication, measurements, and more) to improve the accuracy of its predictions; and its (pre-)training results in disease and patient representations that can help us get a step closer to interpretable predictions.
Learning Multimorbidity Patterns from Electronic Health Records Using Non-negative Matrix Factorisation
Jul 19, 2019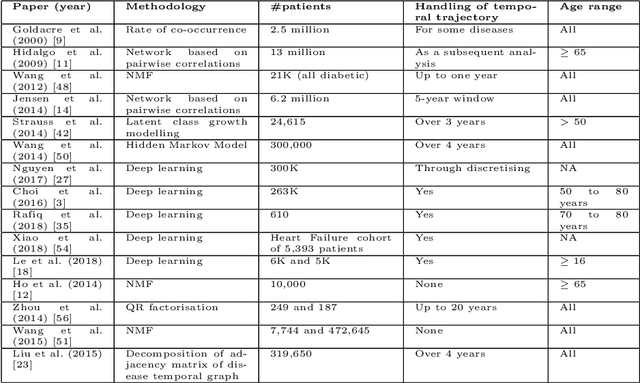
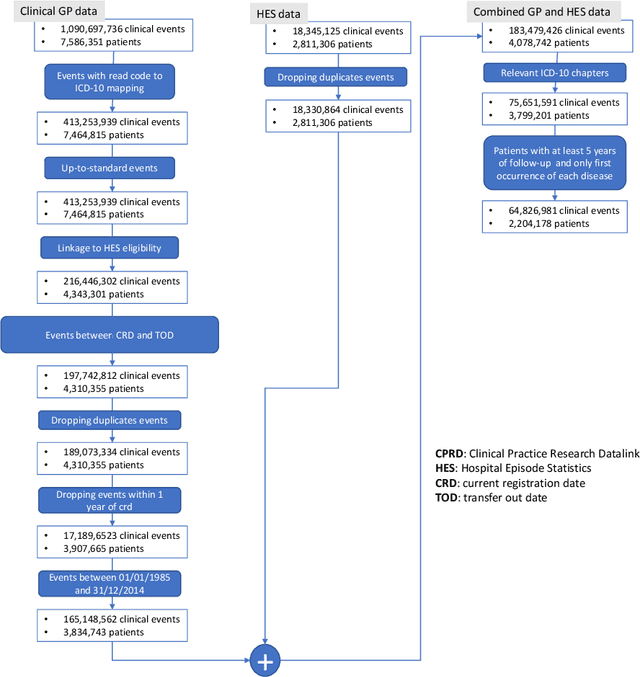
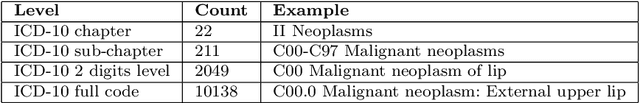
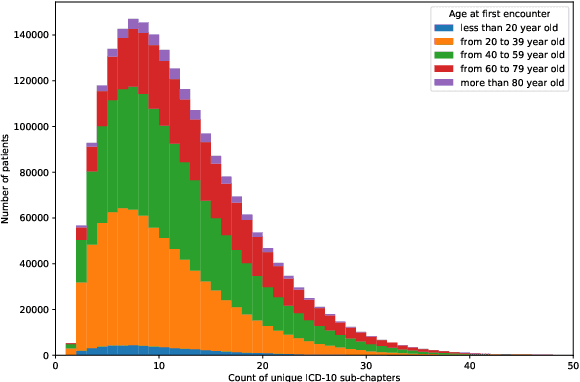
Abstract:Multimorbidity, or the presence of several medical conditions in the same individual, have been increasing in the population both in absolute and relative terms. However, multimorbidity remains poorly understood, and the evidence from existing research to describe its burden, determinants and consequences have been limited. Many of these studies are often cross-sectional and do not explicitly account for multimorbidity patterns' evolution over time. Some studies were based on small datasets, used arbitrary or narrow age range, or lacked appropriate clinical validations. In this study, we applied Non-negative Matrix Factorisation (NMF) in a novel way to one of the largest electronic health records (EHR) databases in the world (with 4 million patients), for simultaneously modelling disease clusters and their role in one's multimorbidity over time. Furthermore, we demonstrated how the temporal characteristics that our model associates with each disease cluster can help mine disease trajectories/networks and generate new hypotheses for the formation of multimorbidity clusters as a function of time/ageing. Our results suggest that our method's ability to learn the underlying dynamics of diseases can provide the field with a novel data-driven / exploratory way of learning the patterns of multimorbidity and their interactions over time.
Performance Measurement for Deep Bayesian Neural Network
Mar 22, 2019
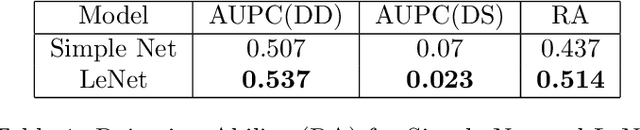
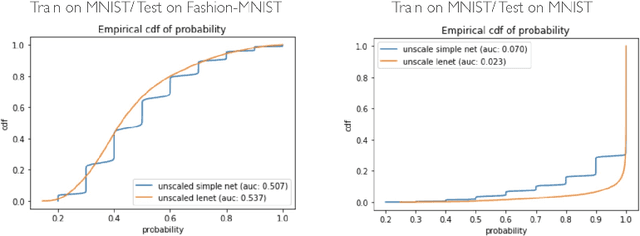
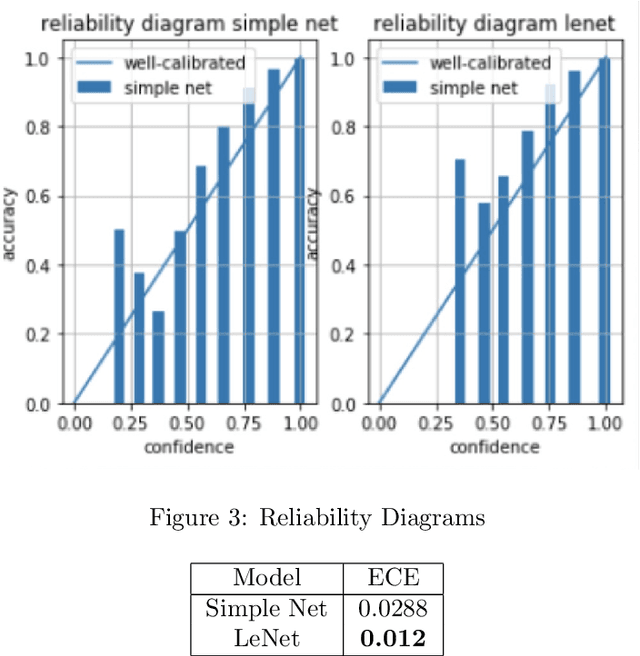
Abstract:Deep Bayesian neural network has aroused a great attention in recent years since it combines the benefits of deep neural network and probability theory. Because of this, the network can make predictions and quantify the uncertainty of the predictions at the same time, which is important in many life-threatening areas. However, most of the recent researches are mainly focusing on making the Bayesian neural network easier to train, and proposing methods to estimate the uncertainty. I notice there are very few works that properly discuss the ways to measure the performance of the Bayesian neural network. Although accuracy and average uncertainty are commonly used for now, they are too general to provide any insight information about the model. In this paper, we would like to introduce more specific criteria and propose several metrics to measure the model performance from different perspectives, which include model calibration measurement, data rejection ability and uncertainty divergence for samples from the same and different distributions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge