Adrienne Kline
YOLO-Angio: An Algorithm for Coronary Anatomy Segmentation
Oct 24, 2023Abstract:Coronary angiography remains the gold standard for diagnosis of coronary artery disease, the most common cause of death worldwide. While this procedure is performed more than 2 million times annually, there remain few methods for fast and accurate automated measurement of disease and localization of coronary anatomy. Here, we present our solution to the Automatic Region-based Coronary Artery Disease diagnostics using X-ray angiography images (ARCADE) challenge held at MICCAI 2023. For the artery segmentation task, our three-stage approach combines preprocessing and feature selection by classical computer vision to enhance vessel contrast, followed by an ensemble model based on YOLOv8 to propose possible vessel candidates by generating a vessel map. A final segmentation is based on a logic-based approach to reconstruct the coronary tree in a graph-based sorting method. Our entry to the ARCADE challenge placed 3rd overall. Using the official metric for evaluation, we achieved an F1 score of 0.422 and 0.4289 on the validation and hold-out sets respectively.
StenUNet: Automatic Stenosis Detection from X-ray Coronary Angiography
Oct 23, 2023



Abstract:Coronary angiography continues to serve as the primary method for diagnosing coronary artery disease (CAD), which is the leading global cause of mortality. The severity of CAD is quantified by the location, degree of narrowing (stenosis), and number of arteries involved. In current practice, this quantification is performed manually using visual inspection and thus suffers from poor inter- and intra-rater reliability. The MICCAI grand challenge: Automatic Region-based Coronary Artery Disease diagnostics using the X-ray angiography imagEs (ARCADE) curated a dataset with stenosis annotations, with the goal of creating an automated stenosis detection algorithm. Using a combination of machine learning and other computer vision techniques, we propose the architecture and algorithm StenUNet to accurately detect stenosis from X-ray Coronary Angiography. Our submission to the ARCADE challenge placed 3rd among all teams. We achieved an F1 score of 0.5348 on the test set, 0.0005 lower than the 2nd place.
Medical Image Deidentification, Cleaning and Compression Using Pylogik
May 03, 2023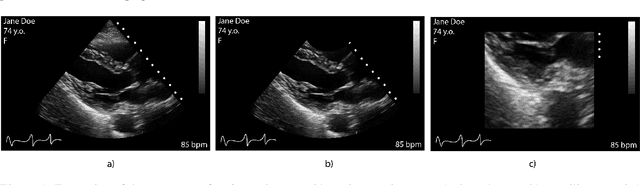
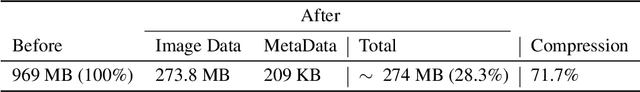
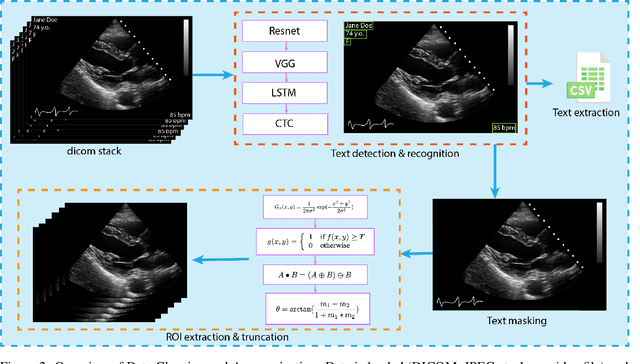
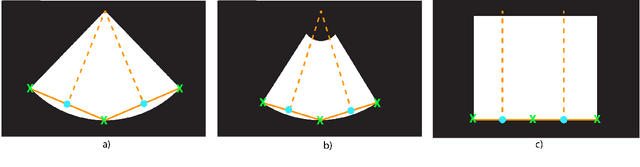
Abstract:Leveraging medical record information in the era of big data and machine learning comes with the caveat that data must be cleaned and deidentified. Facilitating data sharing and harmonization for multi-center collaborations are particularly difficult when protected health information (PHI) is contained or embedded in image meta-data. We propose a novel library in the Python framework, called PyLogik, to help alleviate this issue for ultrasound images, which are particularly challenging because of the frequent inclusion of PHI directly on the images. PyLogik processes the image volumes through a series of text detection/extraction, filtering, thresholding, morphological and contour comparisons. This methodology deidentifies the images, reduces file sizes, and prepares image volumes for applications in deep learning and data sharing. To evaluate its effectiveness in the identification of regions of interest (ROI), a random sample of 50 cardiac ultrasounds (echocardiograms) were processed through PyLogik, and the outputs were compared with the manual segmentations by an expert user. The Dice coefficient of the two approaches achieved an average value of 0.976. Next, an investigation was conducted to ascertain the degree of information compression achieved using the algorithm. Resultant data was found to be on average approximately 72% smaller after processing by PyLogik. Our results suggest that PyLogik is a viable methodology for ultrasound data cleaning and deidentification, determining ROI, and file compression which will facilitate efficient storage, use, and dissemination of ultrasound data.
Machine Learning Capability: A standardized metric using case difficulty with applications to individualized deployment of supervised machine learning
Feb 09, 2023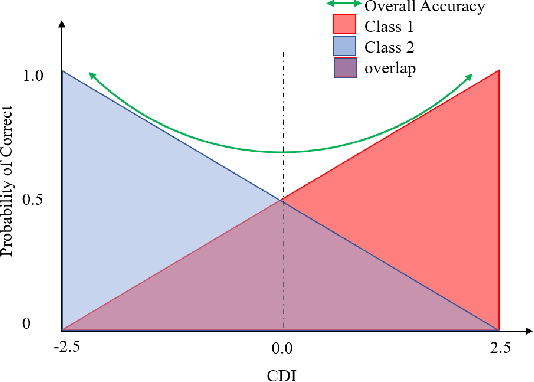
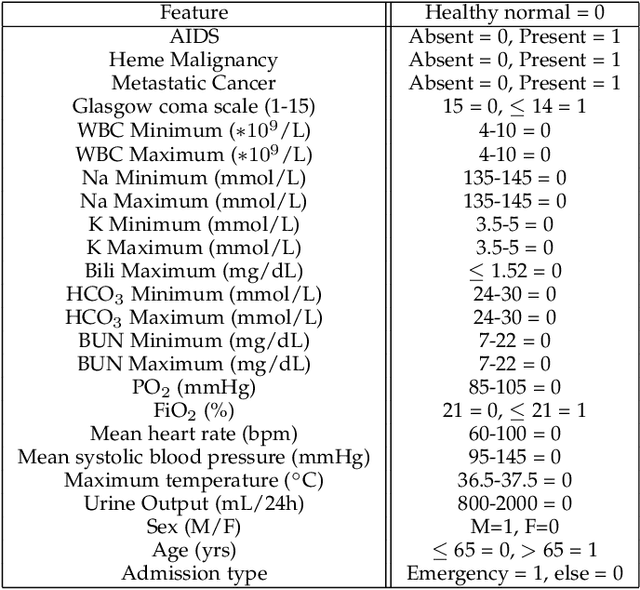
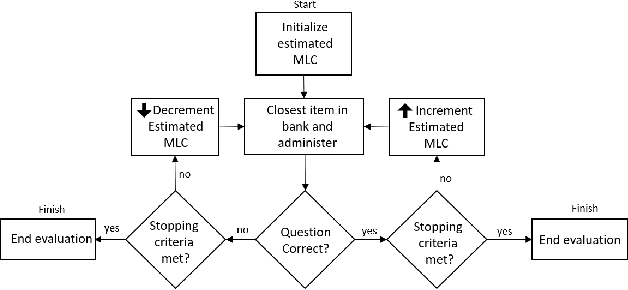
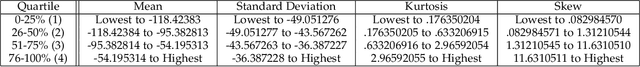
Abstract:Model evaluation is a critical component in supervised machine learning classification analyses. Traditional metrics do not currently incorporate case difficulty. This renders the classification results unbenchmarked for generalization. Item Response Theory (IRT) and Computer Adaptive Testing (CAT) with machine learning can benchmark datasets independent of the end-classification results. This provides high levels of case-level information regarding evaluation utility. To showcase, two datasets were used: 1) health-related and 2) physical science. For the health dataset a two-parameter IRT model, and for the physical science dataset a polytonomous IRT model, was used to analyze predictive features and place each case on a difficulty continuum. A CAT approach was used to ascertain the algorithms' performance and applicability to new data. This method provides an efficient way to benchmark data, using only a fraction of the dataset (less than 1%) and 22-60x more computationally efficient than traditional metrics. This novel metric, termed Machine Learning Capability (MLC) has additional benefits as it is unbiased to outcome classification and a standardized way to make model comparisons within and across datasets. MLC provides a metric on the limitation of supervised machine learning algorithms. In situations where the algorithm falls short, other input(s) are required for decision-making.
IRTCI: Item Response Theory for Categorical Imputation
Feb 08, 2023



Abstract:Most datasets suffer from partial or complete missing values, which has downstream limitations on the available models on which to test the data and on any statistical inferences that can be made from the data. Several imputation techniques have been designed to replace missing data with stand in values. The various approaches have implications for calculating clinical scores, model building and model testing. The work showcased here offers a novel means for categorical imputation based on item response theory (IRT) and compares it against several methodologies currently used in the machine learning field including k-nearest neighbors (kNN), multiple imputed chained equations (MICE) and Amazon Web Services (AWS) deep learning method, Datawig. Analyses comparing these techniques were performed on three different datasets that represented ordinal, nominal and binary categories. The data were modified so that they also varied on both the proportion of data missing and the systematization of the missing data. Two different assessments of performance were conducted: accuracy in reproducing the missing values, and predictive performance using the imputed data. Results demonstrated that the new method, Item Response Theory for Categorical Imputation (IRTCI), fared quite well compared to currently used methods, outperforming several of them in many conditions. Given the theoretical basis for the new approach, and the unique generation of probabilistic terms for determining category belonging for missing cells, IRTCI offers a viable alternative to current approaches.
Multimodal Machine Learning in Precision Health
Apr 10, 2022



Abstract:As machine learning and artificial intelligence are more frequently being leveraged to tackle problems in the health sector, there has been increased interest in utilizing them in clinical decision-support. This has historically been the case in single modal data such as electronic health record data. Attempts to improve prediction and resemble the multimodal nature of clinical expert decision-making this has been met in the computational field of machine learning by a fusion of disparate data. This review was conducted to summarize this field and identify topics ripe for future research. We conducted this review in accordance with the PRISMA (Preferred Reporting Items for Systematic reviews and Meta-Analyses) extension for Scoping Reviews to characterize multi-modal data fusion in health. We used a combination of content analysis and literature searches to establish search strings and databases of PubMed, Google Scholar, and IEEEXplore from 2011 to 2021. A final set of 125 articles were included in the analysis. The most common health areas utilizing multi-modal methods were neurology and oncology. However, there exist a wide breadth of current applications. The most common form of information fusion was early fusion. Notably, there was an improvement in predictive performance performing heterogeneous data fusion. Lacking from the papers were clear clinical deployment strategies and pursuit of FDA-approved tools. These findings provide a map of the current literature on multimodal data fusion as applied to health diagnosis/prognosis problems. Multi-modal machine learning, while more robust in its estimations over unimodal methods, has drawbacks in its scalability and the time-consuming nature of information concatenation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge