Sanjiv Shah
Medical Image Deidentification, Cleaning and Compression Using Pylogik
May 03, 2023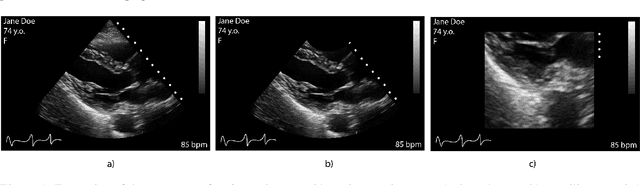
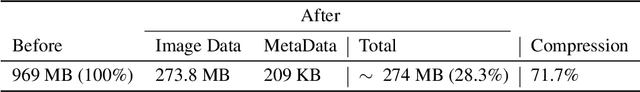
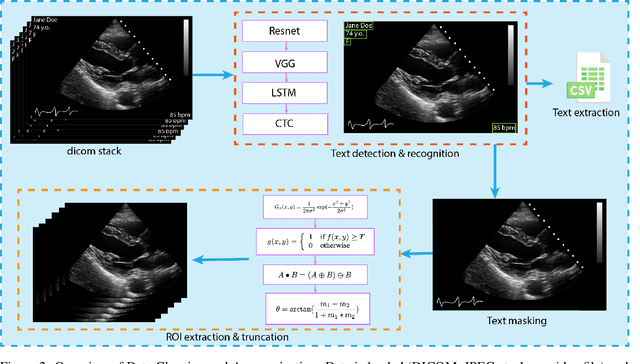
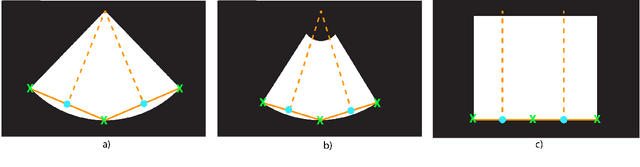
Abstract:Leveraging medical record information in the era of big data and machine learning comes with the caveat that data must be cleaned and deidentified. Facilitating data sharing and harmonization for multi-center collaborations are particularly difficult when protected health information (PHI) is contained or embedded in image meta-data. We propose a novel library in the Python framework, called PyLogik, to help alleviate this issue for ultrasound images, which are particularly challenging because of the frequent inclusion of PHI directly on the images. PyLogik processes the image volumes through a series of text detection/extraction, filtering, thresholding, morphological and contour comparisons. This methodology deidentifies the images, reduces file sizes, and prepares image volumes for applications in deep learning and data sharing. To evaluate its effectiveness in the identification of regions of interest (ROI), a random sample of 50 cardiac ultrasounds (echocardiograms) were processed through PyLogik, and the outputs were compared with the manual segmentations by an expert user. The Dice coefficient of the two approaches achieved an average value of 0.976. Next, an investigation was conducted to ascertain the degree of information compression achieved using the algorithm. Resultant data was found to be on average approximately 72% smaller after processing by PyLogik. Our results suggest that PyLogik is a viable methodology for ultrasound data cleaning and deidentification, determining ROI, and file compression which will facilitate efficient storage, use, and dissemination of ultrasound data.
A Bootstrap Machine Learning Approach to Identify Rare Disease Patients from Electronic Health Records
Sep 06, 2016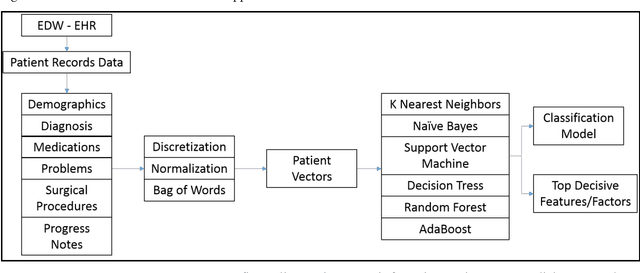

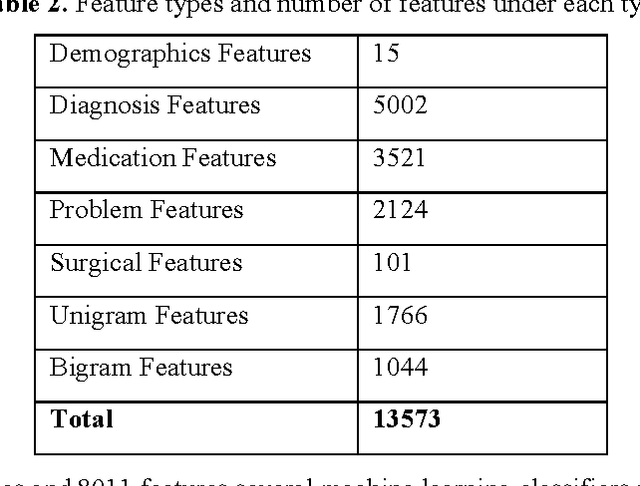

Abstract:Rare diseases are very difficult to identify among large number of other possible diagnoses. Better availability of patient data and improvement in machine learning algorithms empower us to tackle this problem computationally. In this paper, we target one such rare disease - cardiac amyloidosis. We aim to automate the process of identifying potential cardiac amyloidosis patients with the help of machine learning algorithms and also learn most predictive factors. With the help of experienced cardiologists, we prepared a gold standard with 73 positive (cardiac amyloidosis) and 197 negative instances. We achieved high average cross-validation F1 score of 0.98 using an ensemble machine learning classifier. Some of the predictive variables were: Age and Diagnosis of cardiac arrest, chest pain, congestive heart failure, hypertension, prim open angle glaucoma, and shoulder arthritis. Further studies are needed to validate the accuracy of the system across an entire health system and its generalizability for other diseases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge