Chengsheng Mao
Deep Reinforcement Learning for Efficient and Fair Allocation of Health Care Resources
Sep 15, 2023



Abstract:Scarcity of health care resources could result in the unavoidable consequence of rationing. For example, ventilators are often limited in supply, especially during public health emergencies or in resource-constrained health care settings, such as amid the pandemic of COVID-19. Currently, there is no universally accepted standard for health care resource allocation protocols, resulting in different governments prioritizing patients based on various criteria and heuristic-based protocols. In this study, we investigate the use of reinforcement learning for critical care resource allocation policy optimization to fairly and effectively ration resources. We propose a transformer-based deep Q-network to integrate the disease progression of individual patients and the interaction effects among patients during the critical care resource allocation. We aim to improve both fairness of allocation and overall patient outcomes. Our experiments demonstrate that our method significantly reduces excess deaths and achieves a more equitable distribution under different levels of ventilator shortage, when compared to existing severity-based and comorbidity-based methods in use by different governments. Our source code is included in the supplement and will be released on Github upon publication.
Exploring Large Language Models for Knowledge Graph Completion
Sep 10, 2023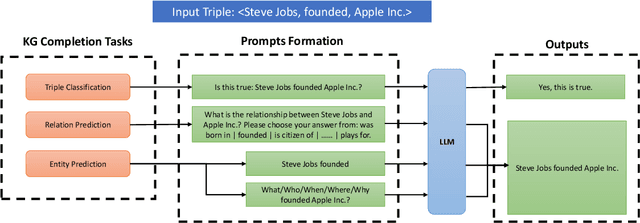
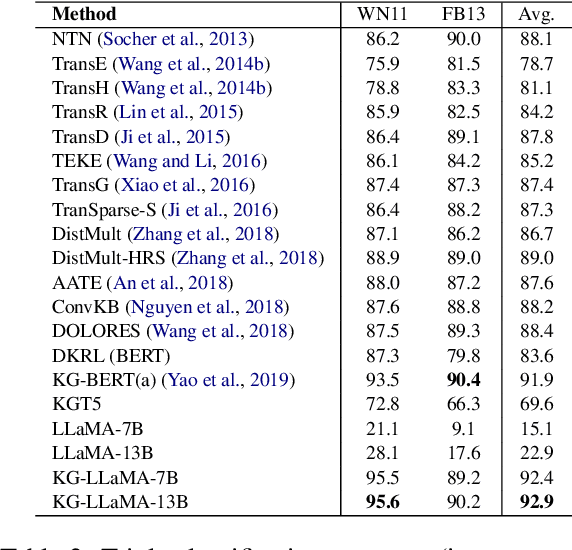
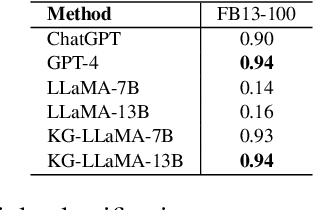
Abstract:Knowledge graphs play a vital role in numerous artificial intelligence tasks, yet they frequently face the issue of incompleteness. In this study, we explore utilizing Large Language Models (LLM) for knowledge graph completion. We consider triples in knowledge graphs as text sequences and introduce an innovative framework called Knowledge Graph LLM (KG-LLM) to model these triples. Our technique employs entity and relation descriptions of a triple as prompts and utilizes the response for predictions. Experiments on various benchmark knowledge graphs demonstrate that our method attains state-of-the-art performance in tasks such as triple classification and relation prediction. We also find that fine-tuning relatively smaller models (e.g., LLaMA-7B, ChatGLM-6B) outperforms recent ChatGPT and GPT-4.
AKI-BERT: a Pre-trained Clinical Language Model for Early Prediction of Acute Kidney Injury
May 07, 2022



Abstract:Acute kidney injury (AKI) is a common clinical syndrome characterized by a sudden episode of kidney failure or kidney damage within a few hours or a few days. Accurate early prediction of AKI for patients in ICU who are more likely than others to have AKI can enable timely interventions, and reduce the complications of AKI. Much of the clinical information relevant to AKI is captured in clinical notes that are largely unstructured text and requires advanced natural language processing (NLP) for useful information extraction. On the other hand, pre-trained contextual language models such as Bidirectional Encoder Representations from Transformers (BERT) have improved performances for many NLP tasks in general domain recently. However, few have explored BERT on disease-specific medical domain tasks such as AKI early prediction. In this paper, we try to apply BERT to specific diseases and present an AKI domain-specific pre-trained language model based on BERT (AKI-BERT) that could be used to mine the clinical notes for early prediction of AKI. AKI-BERT is a BERT model pre-trained on the clinical notes of patients having risks for AKI. Our experiments on Medical Information Mart for Intensive Care III (MIMIC-III) dataset demonstrate that AKI-BERT can yield performance improvements for early AKI prediction, thus expanding the utility of the BERT model from general clinical domain to disease-specific domain.
Distribution Preserving Graph Representation Learning
Feb 27, 2022



Abstract:Graph neural network (GNN) is effective to model graphs for distributed representations of nodes and an entire graph. Recently, research on the expressive power of GNN attracted growing attention. A highly-expressive GNN has the ability to generate discriminative graph representations. However, in the end-to-end training process for a certain graph learning task, a highly-expressive GNN risks generating graph representations overfitting the training data for the target task, while losing information important for the model generalization. In this paper, we propose Distribution Preserving GNN (DP-GNN) - a GNN framework that can improve the generalizability of expressive GNN models by preserving several kinds of distribution information in graph representations and node representations. Besides the generalizability, by applying an expressive GNN backbone, DP-GNN can also have high expressive power. We evaluate the proposed DP-GNN framework on multiple benchmark datasets for graph classification tasks. The experimental results demonstrate that our model achieves state-of-the-art performances.
Constrained tensor factorization for computational phenotyping and mortality prediction in patients with cancer
Dec 24, 2021



Abstract:Background: The increasing adoption of electronic health records (EHR) across the US has created troves of computable data, to which machine learning methods have been applied to extract useful insights. EHR data, represented as a three-dimensional analogue of a matrix (tensor), is decomposed into two-dimensional factors that can be interpreted as computational phenotypes. Methods: We apply constrained tensor factorization to derive computational phenotypes and predict mortality in cohorts of patients with breast, prostate, colorectal, or lung cancer in the Northwestern Medicine Enterprise Data Warehouse from 2000 to 2015. In our experiments, we examined using a supervised term in the factorization algorithm, filtering tensor co-occurrences by medical indication, and incorporating additional social determinants of health (SDOH) covariates in the factorization process. We evaluated the resulting computational phenotypes qualitatively and by assessing their ability to predict five-year mortality using the area under the curve (AUC) statistic. Results: Filtering by medical indication led to more concise and interpretable phenotypes. Mortality prediction performance (AUC) varied under the different experimental conditions and by cancer type (breast: 0.623 - 0.694, prostate: 0.603 - 0.750, colorectal: 0.523 - 0.641, and lung: 0.517 - 0.623). Generally, prediction performance improved with the use of a supervised term and the incorporation of SDOH covariates. Conclusion: Constrained tensor factorization, applied to sparse EHR data of patients with cancer, can discover computational phenotypes predictive of five-year mortality. The incorporation of SDOH variables into the factorization algorithm is an easy-to-implement and effective way to improve prediction performance.
Natural language processing to identify lupus nephritis phenotype in electronic health records
Dec 20, 2021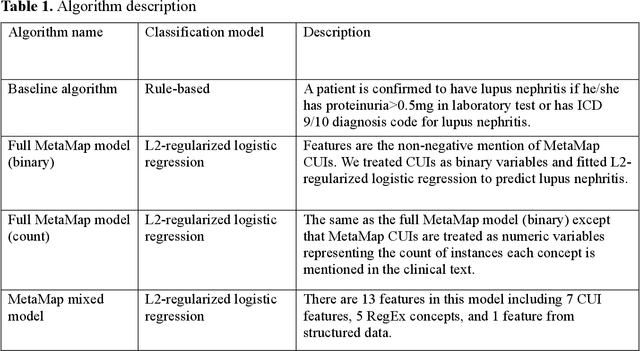
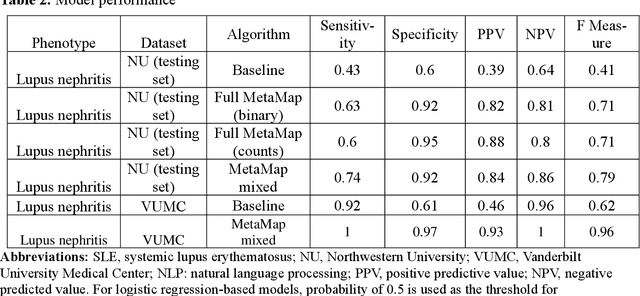
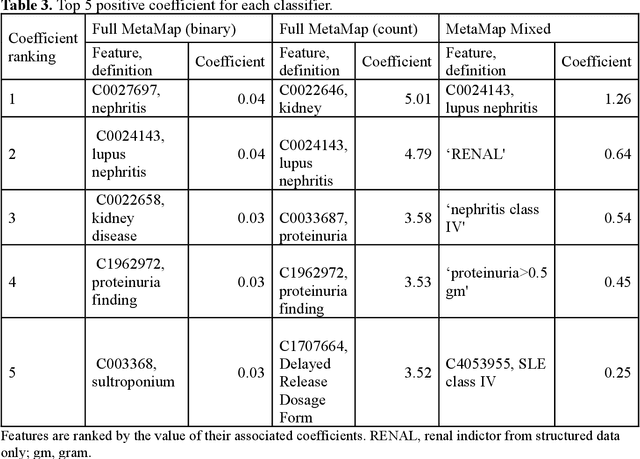
Abstract:Systemic lupus erythematosus (SLE) is a rare autoimmune disorder characterized by an unpredictable course of flares and remission with diverse manifestations. Lupus nephritis, one of the major disease manifestations of SLE for organ damage and mortality, is a key component of lupus classification criteria. Accurately identifying lupus nephritis in electronic health records (EHRs) would therefore benefit large cohort observational studies and clinical trials where characterization of the patient population is critical for recruitment, study design, and analysis. Lupus nephritis can be recognized through procedure codes and structured data, such as laboratory tests. However, other critical information documenting lupus nephritis, such as histologic reports from kidney biopsies and prior medical history narratives, require sophisticated text processing to mine information from pathology reports and clinical notes. In this study, we developed algorithms to identify lupus nephritis with and without natural language processing (NLP) using EHR data. We developed four algorithms: a rule-based algorithm using only structured data (baseline algorithm) and three algorithms using different NLP models. The three NLP models are based on regularized logistic regression and use different sets of features including positive mention of concept unique identifiers (CUIs), number of appearances of CUIs, and a mixture of three components respectively. The baseline algorithm and the best performed NLP algorithm were external validated on a dataset from Vanderbilt University Medical Center (VUMC). Our best performing NLP model incorporating features from both structured data, regular expression concepts, and mapped CUIs improved F measure in both the NMEDW (0.41 vs 0.79) and VUMC (0.62 vs 0.96) datasets compared to the baseline lupus nephritis algorithm.
PANTHER: Pathway Augmented Nonnegative Tensor factorization for HighER-order feature learning
Dec 15, 2020



Abstract:Genetic pathways usually encode molecular mechanisms that can inform targeted interventions. It is often challenging for existing machine learning approaches to jointly model genetic pathways (higher-order features) and variants (atomic features), and present to clinicians interpretable models. In order to build more accurate and better interpretable machine learning models for genetic medicine, we introduce Pathway Augmented Nonnegative Tensor factorization for HighER-order feature learning (PANTHER). PANTHER selects informative genetic pathways that directly encode molecular mechanisms. We apply genetically motivated constrained tensor factorization to group pathways in a way that reflects molecular mechanism interactions. We then train a softmax classifier for disease types using the identified pathway groups. We evaluated PANTHER against multiple state-of-the-art constrained tensor/matrix factorization models, as well as group guided and Bayesian hierarchical models. PANTHER outperforms all state-of-the-art comparison models significantly (p<0.05). Our experiments on large scale Next Generation Sequencing (NGS) and whole-genome genotyping datasets also demonstrated wide applicability of PANTHER. We performed feature analysis in predicting disease types, which suggested insights and benefits of the identified pathway groups.
Towards Expressive Graph Representation
Oct 12, 2020



Abstract:Graph Neural Network (GNN) aggregates the neighborhood of each node into the node embedding and shows its powerful capability for graph representation learning. However, most existing GNN variants aggregate the neighborhood information in a fixed non-injective fashion, which may map different graphs or nodes to the same embedding, reducing the model expressiveness. We present a theoretical framework to design a continuous injective set function for neighborhood aggregation in GNN. Using the framework, we propose expressive GNN that aggregates the neighborhood of each node with a continuous injective set function, so that a GNN layer maps similar nodes with similar neighborhoods to similar embeddings, different nodes to different embeddings and the equivalent nodes or isomorphic graphs to the same embeddings. Moreover, the proposed expressive GNN can naturally learn expressive representations for graphs with continuous node attributes. We validate the proposed expressive GNN (ExpGNN) for graph classification on multiple benchmark datasets including simple graphs and attributed graphs. The experimental results demonstrate that our model achieves state-of-the-art performances on most of the benchmarks.
KG-BERT: BERT for Knowledge Graph Completion
Sep 11, 2019



Abstract:Knowledge graphs are important resources for many artificial intelligence tasks but often suffer from incompleteness. In this work, we propose to use pre-trained language models for knowledge graph completion. We treat triples in knowledge graphs as textual sequences and propose a novel framework named Knowledge Graph Bidirectional Encoder Representations from Transformer (KG-BERT) to model these triples. Our method takes entity and relation descriptions of a triple as input and computes scoring function of the triple with the KG-BERT language model. Experimental results on multiple benchmark knowledge graphs show that our method can achieve state-of-the-art performance in triple classification, link prediction and relation prediction tasks.
MedGCN: Graph Convolutional Networks for Multiple Medical Tasks
Mar 31, 2019



Abstract:Laboratory testing and medication prescription are two of the most important routines in daily clinical practice. Developing an artificial intelligence system that can automatically make lab test imputations and medication recommendations can save cost on potentially redundant lab tests and inform physicians in more effective prescription. We present an intelligent model that can automatically recommend the patients' medications based on their incomplete lab tests, and can even accurately estimate the lab values that have not been taken. We model the complex relations between multiple types of medical entities with their inherent features in a heterogeneous graph. Then we learn a distributed representation for each entity in the graph based on graph convolutional networks to make the representations integrate information from multiple types of entities. Since the entity representations incorporate multiple types of medical information, they can be used for multiple medical tasks. In our experiments, we construct a graph to associate patients, encounters, lab tests and medications, and conduct the two tasks: medication recommendation and lab test imputation. The experimental results demonstrate that our model can outperform the state-of-the-art models in both tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge