Zhenbang Wu
PyHealth 2.0: A Comprehensive Open-Source Toolkit for Accessible and Reproducible Clinical Deep Learning
Jan 23, 2026Abstract:Difficulty replicating baselines, high computational costs, and required domain expertise create persistent barriers to clinical AI research. To address these challenges, we introduce PyHealth 2.0, an enhanced clinical deep learning toolkit that enables predictive modeling in as few as 7 lines of code. PyHealth 2.0 offers three key contributions: (1) a comprehensive toolkit addressing reproducibility and compatibility challenges by unifying 15+ datasets, 20+ clinical tasks, 25+ models, 5+ interpretability methods, and uncertainty quantification including conformal prediction within a single framework that supports diverse clinical data modalities - signals, imaging, and electronic health records - with translation of 5+ medical coding standards; (2) accessibility-focused design accommodating multimodal data and diverse computational resources with up to 39x faster processing and 20x lower memory usage, enabling work from 16GB laptops to production systems; and (3) an active open-source community of 400+ members lowering domain expertise barriers through extensive documentation, reproducible research contributions, and collaborations with academic health systems and industry partners, including multi-language support via RHealth. PyHealth 2.0 establishes an open-source foundation and community advancing accessible, reproducible healthcare AI. Available at pip install pyhealth.
Training LLMs for EHR-Based Reasoning Tasks via Reinforcement Learning
May 30, 2025Abstract:We present EHRMIND, a practical recipe for adapting large language models (LLMs) to complex clinical reasoning tasks using reinforcement learning with verifiable rewards (RLVR). While RLVR has succeeded in mathematics and coding, its application to healthcare contexts presents unique challenges due to the specialized knowledge and reasoning required for electronic health record (EHR) interpretation. Our pilot study on the MEDCALC benchmark reveals two key failure modes: (1) misapplied knowledge, where models possess relevant medical knowledge but apply it incorrectly, and (2) missing knowledge, where models lack essential domain knowledge. To address these cases, EHRMIND applies a two-stage solution: a lightweight supervised fine-tuning (SFT) warm-up that injects missing domain knowledge, stabilizes subsequent training, and encourages structured, interpretable outputs; followed by RLVR, which reinforces outcome correctness and refines the model's decision-making. We demonstrate the effectiveness of our method across diverse clinical applications, including medical calculations (MEDCALC), patient-trial matching (TREC CLINICAL TRIALS), and disease diagnosis (EHRSHOT). EHRMIND delivers consistent gains in accuracy, interpretability, and cross-task generalization. These findings offer practical guidance for applying RLVR to enhance LLM capabilities in healthcare settings.
Reinforcement Learning for Out-of-Distribution Reasoning in LLMs: An Empirical Study on Diagnosis-Related Group Coding
May 28, 2025Abstract:Diagnosis-Related Group (DRG) codes are essential for hospital reimbursement and operations but require labor-intensive assignment. Large Language Models (LLMs) struggle with DRG coding due to the out-of-distribution (OOD) nature of the task: pretraining corpora rarely contain private clinical or billing data. We introduce DRG-Sapphire, which uses large-scale reinforcement learning (RL) for automated DRG coding from clinical notes. Built on Qwen2.5-7B and trained with Group Relative Policy Optimization (GRPO) using rule-based rewards, DRG-Sapphire introduces a series of RL enhancements to address domain-specific challenges not seen in previous mathematical tasks. Our model achieves state-of-the-art accuracy on the MIMIC-IV benchmark and generates physician-validated reasoning for DRG assignments, significantly enhancing explainability. Our study further sheds light on broader challenges of applying RL to knowledge-intensive, OOD tasks. We observe that RL performance scales approximately linearly with the logarithm of the number of supervised fine-tuning (SFT) examples, suggesting that RL effectiveness is fundamentally constrained by the domain knowledge encoded in the base model. For OOD tasks like DRG coding, strong RL performance requires sufficient knowledge infusion prior to RL. Consequently, scaling SFT may be more effective and computationally efficient than scaling RL alone for such tasks.
CARES: A Comprehensive Benchmark of Trustworthiness in Medical Vision Language Models
Jun 10, 2024
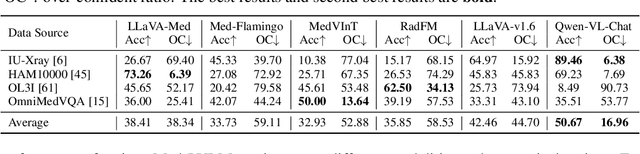
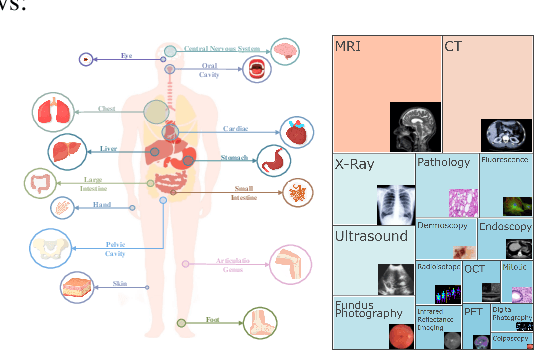
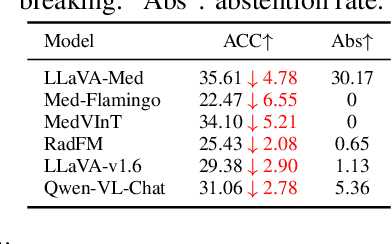
Abstract:Artificial intelligence has significantly impacted medical applications, particularly with the advent of Medical Large Vision Language Models (Med-LVLMs), sparking optimism for the future of automated and personalized healthcare. However, the trustworthiness of Med-LVLMs remains unverified, posing significant risks for future model deployment. In this paper, we introduce CARES and aim to comprehensively evaluate the Trustworthiness of Med-LVLMs across the medical domain. We assess the trustworthiness of Med-LVLMs across five dimensions, including trustfulness, fairness, safety, privacy, and robustness. CARES comprises about 41K question-answer pairs in both closed and open-ended formats, covering 16 medical image modalities and 27 anatomical regions. Our analysis reveals that the models consistently exhibit concerns regarding trustworthiness, often displaying factual inaccuracies and failing to maintain fairness across different demographic groups. Furthermore, they are vulnerable to attacks and demonstrate a lack of privacy awareness. We publicly release our benchmark and code in https://github.com/richard-peng-xia/CARES.
MedCLIP: Contrastive Learning from Unpaired Medical Images and Text
Oct 18, 2022



Abstract:Existing vision-text contrastive learning like CLIP aims to match the paired image and caption embeddings while pushing others apart, which improves representation transferability and supports zero-shot prediction. However, medical image-text datasets are orders of magnitude below the general images and captions from the internet. Moreover, previous methods encounter many false negatives, i.e., images and reports from separate patients probably carry the same semantics but are wrongly treated as negatives. In this paper, we decouple images and texts for multimodal contrastive learning thus scaling the usable training data in a combinatorial magnitude with low cost. We also propose to replace the InfoNCE loss with semantic matching loss based on medical knowledge to eliminate false negatives in contrastive learning. We prove that MedCLIP is a simple yet effective framework: it outperforms state-of-the-art methods on zero-shot prediction, supervised classification, and image-text retrieval. Surprisingly, we observe that with only 20K pre-training data, MedCLIP wins over the state-of-the-art method (using around 200K data). Our code is available at https://github.com/RyanWangZf/MedCLIP.
Knowledge-Driven New Drug Recommendation
Oct 11, 2022



Abstract:Drug recommendation assists doctors in prescribing personalized medications to patients based on their health conditions. Existing drug recommendation solutions adopt the supervised multi-label classification setup and only work with existing drugs with sufficient prescription data from many patients. However, newly approved drugs do not have much historical prescription data and cannot leverage existing drug recommendation methods. To address this, we formulate the new drug recommendation as a few-shot learning problem. Yet, directly applying existing few-shot learning algorithms faces two challenges: (1) complex relations among diseases and drugs and (2) numerous false-negative patients who were eligible but did not yet use the new drugs. To tackle these challenges, we propose EDGE, which can quickly adapt to the recommendation for a new drug with limited prescription data from a few support patients. EDGE maintains a drug-dependent multi-phenotype few-shot learner to bridge the gap between existing and new drugs. Specifically, EDGE leverages the drug ontology to link new drugs to existing drugs with similar treatment effects and learns ontology-based drug representations. Such drug representations are used to customize the metric space of the phenotype-driven patient representations, which are composed of a set of phenotypes capturing complex patient health status. Lastly, EDGE eliminates the false-negative supervision signal using an external drug-disease knowledge base. We evaluate EDGE on two real-world datasets: the public EHR data (MIMIC-IV) and private industrial claims data. Results show that EDGE achieves 7.3% improvement on the ROC-AUC score over the best baseline.
AutoMap: Automatic Medical Code Mapping for Clinical Prediction Model Deployment
Mar 04, 2022



Abstract:Given a deep learning model trained on data from a source site, how to deploy the model to a target hospital automatically? How to accommodate heterogeneous medical coding systems across different hospitals? Standard approaches rely on existing medical code mapping tools, which have significant practical limitations. To tackle this problem, we propose AutoMap to automatically map the medical codes across different EHR systems in a coarse-to-fine manner: (1) Ontology-level Alignment: We leverage the ontology structure to learn a coarse alignment between the source and target medical coding systems; (2) Code-level Refinement: We refine the alignment at a fine-grained code level for the downstream tasks using a teacher-student framework. We evaluate AutoMap using several deep learning models with two real-world EHR datasets: eICU and MIMIC-III. Results show that AutoMap achieves relative improvements up to 3.9% (AUC-ROC) and 8.7% (AUC-PR) for mortality prediction, and up to 4.7% (AUC-ROC) and 3.7% (F1) for length-of-stay estimation. Further, we show that AutoMap can provide accurate mapping across coding systems. Lastly, we demonstrate that AutoMap can adapt to the two challenging scenarios: (1) mapping between completely different coding systems and (2) between completely different hospitals.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge