Zifeng Wang
SAGE: Steerable Agentic Data Generation for Deep Search with Execution Feedback
Jan 26, 2026Abstract:Deep search agents, which aim to answer complex questions requiring reasoning across multiple documents, can significantly speed up the information-seeking process. Collecting human annotations for this application is prohibitively expensive due to long and complex exploration trajectories. We propose an agentic pipeline that automatically generates high quality, difficulty-controlled deep search question-answer pairs for a given corpus and a target difficulty level. Our pipeline, SAGE, consists of a data generator which proposes QA pairs and a search agent which attempts to solve the generated question and provide execution feedback for the data generator. The two components interact over multiple rounds to iteratively refine the question-answer pairs until they satisfy the target difficulty level. Our intrinsic evaluation shows SAGE generates questions that require diverse reasoning strategies, while significantly increases the correctness and difficulty of the generated data. Our extrinsic evaluation demonstrates up to 23% relative performance gain on popular deep search benchmarks by training deep search agents with our synthetic data. Additional experiments show that agents trained on our data can adapt from fixed-corpus retrieval to Google Search at inference time, without further training.
Adaptation of Agentic AI
Dec 22, 2025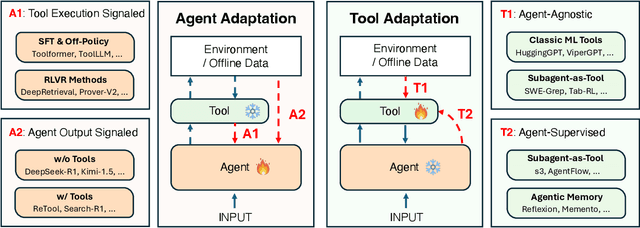
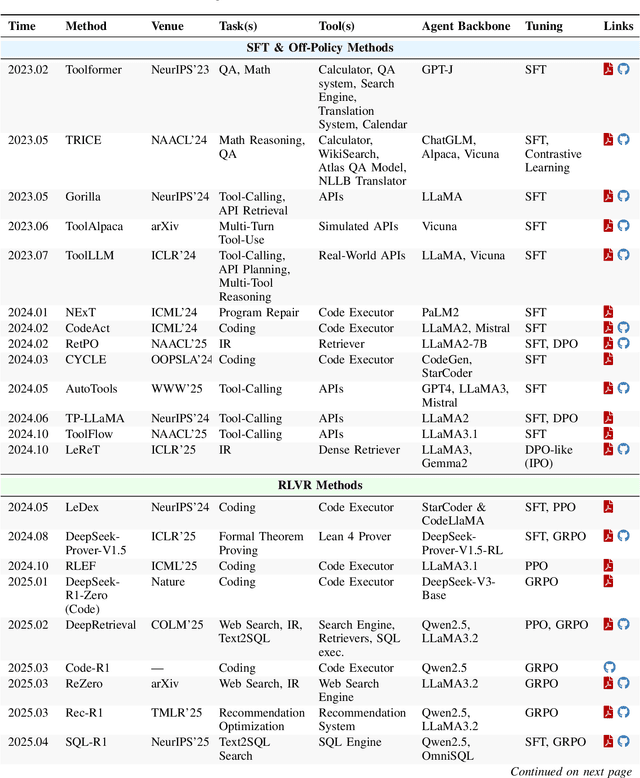
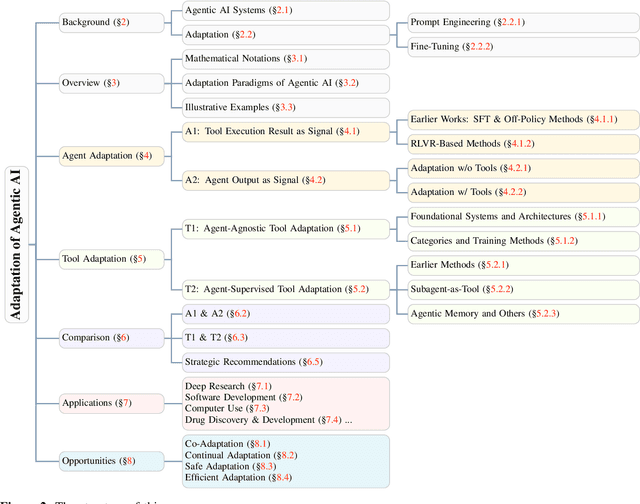
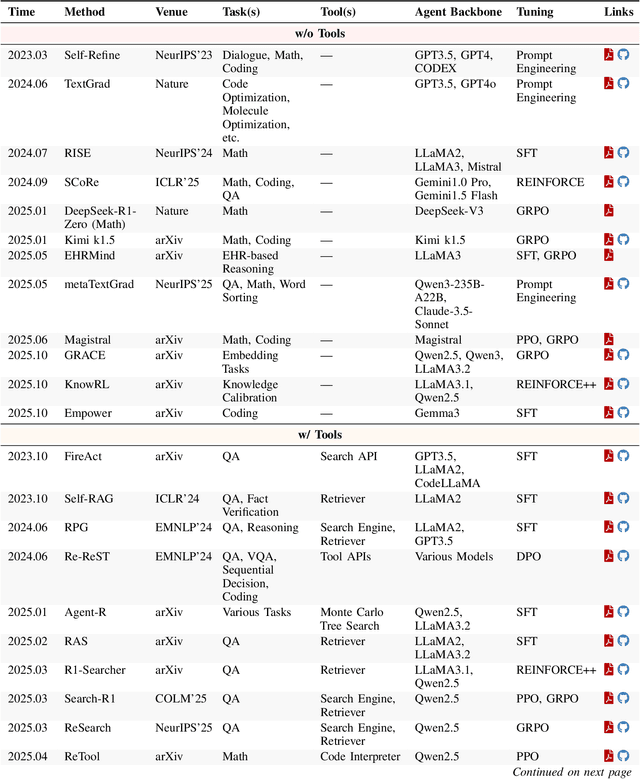
Abstract:Cutting-edge agentic AI systems are built on foundation models that can be adapted to plan, reason, and interact with external tools to perform increasingly complex and specialized tasks. As these systems grow in capability and scope, adaptation becomes a central mechanism for improving performance, reliability, and generalization. In this paper, we unify the rapidly expanding research landscape into a systematic framework that spans both agent adaptations and tool adaptations. We further decompose these into tool-execution-signaled and agent-output-signaled forms of agent adaptation, as well as agent-agnostic and agent-supervised forms of tool adaptation. We demonstrate that this framework helps clarify the design space of adaptation strategies in agentic AI, makes their trade-offs explicit, and provides practical guidance for selecting or switching among strategies during system design. We then review the representative approaches in each category, analyze their strengths and limitations, and highlight key open challenges and future opportunities. Overall, this paper aims to offer a conceptual foundation and practical roadmap for researchers and practitioners seeking to build more capable, efficient, and reliable agentic AI systems.
Supervised Reinforcement Learning: From Expert Trajectories to Step-wise Reasoning
Oct 29, 2025Abstract:Large Language Models (LLMs) often struggle with problems that require multi-step reasoning. For small-scale open-source models, Reinforcement Learning with Verifiable Rewards (RLVR) fails when correct solutions are rarely sampled even after many attempts, while Supervised Fine-Tuning (SFT) tends to overfit long demonstrations through rigid token-by-token imitation. To address this gap, we propose Supervised Reinforcement Learning (SRL), a framework that reformulates problem solving as generating a sequence of logical "actions". SRL trains the model to generate an internal reasoning monologue before committing to each action. It provides smoother rewards based on the similarity between the model's actions and expert actions extracted from the SFT dataset in a step-wise manner. This supervision offers richer learning signals even when all rollouts are incorrect, while encouraging flexible reasoning guided by expert demonstrations. As a result, SRL enables small models to learn challenging problems previously unlearnable by SFT or RLVR. Moreover, initializing training with SRL before refining with RLVR yields the strongest overall performance. Beyond reasoning benchmarks, SRL generalizes effectively to agentic software engineering tasks, establishing it as a robust and versatile training framework for reasoning-oriented LLMs.
Enabling Flexible Multi-LLM Integration for Scalable Knowledge Aggregation
May 28, 2025



Abstract:Large language models (LLMs) have shown remarkable promise but remain challenging to continually improve through traditional finetuning, particularly when integrating capabilities from other specialized LLMs. Popular methods like ensemble and weight merging require substantial memory and struggle to adapt to changing data environments. Recent efforts have transferred knowledge from multiple LLMs into a single target model; however, they suffer from interference and degraded performance among tasks, largely due to limited flexibility in candidate selection and training pipelines. To address these issues, we propose a framework that adaptively selects and aggregates knowledge from diverse LLMs to build a single, stronger model, avoiding the high memory overhead of ensemble and inflexible weight merging. Specifically, we design an adaptive selection network that identifies the most relevant source LLMs based on their scores, thereby reducing knowledge interference. We further propose a dynamic weighted fusion strategy that accounts for the inherent strengths of candidate LLMs, along with a feedback-driven loss function that prevents the selector from converging on a single subset of sources. Experimental results demonstrate that our method can enable a more stable and scalable knowledge aggregation process while reducing knowledge interference by up to 50% compared to existing approaches. Code is avaliable at https://github.com/ZLKong/LLM_Integration
TrialPanorama: Database and Benchmark for Systematic Review and Design of Clinical Trials
May 22, 2025Abstract:Developing artificial intelligence (AI) for vertical domains requires a solid data foundation for both training and evaluation. In this work, we introduce TrialPanorama, a large-scale, structured database comprising 1,657,476 clinical trial records aggregated from 15 global sources. The database captures key aspects of trial design and execution, including trial setups, interventions, conditions, biomarkers, and outcomes, and links them to standard biomedical ontologies such as DrugBank and MedDRA. This structured and ontology-grounded design enables TrialPanorama to serve as a unified, extensible resource for a wide range of clinical trial tasks, including trial planning, design, and summarization. To demonstrate its utility, we derive a suite of benchmark tasks directly from the TrialPanorama database. The benchmark spans eight tasks across two categories: three for systematic review (study search, study screening, and evidence summarization) and five for trial design (arm design, eligibility criteria, endpoint selection, sample size estimation, and trial completion assessment). The experiments using five state-of-the-art large language models (LLMs) show that while general-purpose LLMs exhibit some zero-shot capability, their performance is still inadequate for high-stakes clinical trial workflows. We release TrialPanorama database and the benchmark to facilitate further research on AI for clinical trials.
BioDSA-1K: Benchmarking Data Science Agents for Biomedical Research
May 22, 2025Abstract:Validating scientific hypotheses is a central challenge in biomedical research, and remains difficult for artificial intelligence (AI) agents due to the complexity of real-world data analysis and evidence interpretation. In this work, we present BioDSA-1K, a benchmark designed to evaluate AI agents on realistic, data-driven biomedical hypothesis validation tasks. BioDSA-1K consists of 1,029 hypothesis-centric tasks paired with 1,177 analysis plans, curated from over 300 published biomedical studies to reflect the structure and reasoning found in authentic research workflows. Each task includes a structured hypothesis derived from the original study's conclusions, expressed in the affirmative to reflect the language of scientific reporting, and one or more pieces of supporting evidence grounded in empirical data tables. While these hypotheses mirror published claims, they remain testable using standard statistical or machine learning methods. The benchmark enables evaluation along four axes: (1) hypothesis decision accuracy, (2) alignment between evidence and conclusion, (3) correctness of the reasoning process, and (4) executability of the AI-generated analysis code. Importantly, BioDSA-1K includes non-verifiable hypotheses: cases where the available data are insufficient to support or refute a claim, reflecting a common yet underexplored scenario in real-world science. We propose BioDSA-1K as a foundation for building and evaluating generalizable, trustworthy AI agents for biomedical discovery.
s3: You Don't Need That Much Data to Train a Search Agent via RL
May 20, 2025Abstract:Retrieval-augmented generation (RAG) systems empower large language models (LLMs) to access external knowledge during inference. Recent advances have enabled LLMs to act as search agents via reinforcement learning (RL), improving information acquisition through multi-turn interactions with retrieval engines. However, existing approaches either optimize retrieval using search-only metrics (e.g., NDCG) that ignore downstream utility or fine-tune the entire LLM to jointly reason and retrieve-entangling retrieval with generation and limiting the real search utility and compatibility with frozen or proprietary models. In this work, we propose s3, a lightweight, model-agnostic framework that decouples the searcher from the generator and trains the searcher using a Gain Beyond RAG reward: the improvement in generation accuracy over naive RAG. s3 requires only 2.4k training samples to outperform baselines trained on over 70x more data, consistently delivering stronger downstream performance across six general QA and five medical QA benchmarks.
InformGen: An AI Copilot for Accurate and Compliant Clinical Research Consent Document Generation
Apr 01, 2025Abstract:Leveraging large language models (LLMs) to generate high-stakes documents, such as informed consent forms (ICFs), remains a significant challenge due to the extreme need for regulatory compliance and factual accuracy. Here, we present InformGen, an LLM-driven copilot for accurate and compliant ICF drafting by optimized knowledge document parsing and content generation, with humans in the loop. We further construct a benchmark dataset comprising protocols and ICFs from 900 clinical trials. Experimental results demonstrate that InformGen achieves near 100% compliance with 18 core regulatory rules derived from FDA guidelines, outperforming a vanilla GPT-4o model by up to 30%. Additionally, a user study with five annotators shows that InformGen, when integrated with manual intervention, attains over 90% factual accuracy, significantly surpassing the vanilla GPT-4o model's 57%-82%. Crucially, InformGen ensures traceability by providing inline citations to source protocols, enabling easy verification and maintaining the highest standards of factual integrity.
In Prospect and Retrospect: Reflective Memory Management for Long-term Personalized Dialogue Agents
Mar 11, 2025Abstract:Large Language Models (LLMs) have made significant progress in open-ended dialogue, yet their inability to retain and retrieve relevant information from long-term interactions limits their effectiveness in applications requiring sustained personalization. External memory mechanisms have been proposed to address this limitation, enabling LLMs to maintain conversational continuity. However, existing approaches struggle with two key challenges. First, rigid memory granularity fails to capture the natural semantic structure of conversations, leading to fragmented and incomplete representations. Second, fixed retrieval mechanisms cannot adapt to diverse dialogue contexts and user interaction patterns. In this work, we propose Reflective Memory Management (RMM), a novel mechanism for long-term dialogue agents, integrating forward- and backward-looking reflections: (1) Prospective Reflection, which dynamically summarizes interactions across granularities-utterances, turns, and sessions-into a personalized memory bank for effective future retrieval, and (2) Retrospective Reflection, which iteratively refines the retrieval in an online reinforcement learning (RL) manner based on LLMs' cited evidence. Experiments show that RMM demonstrates consistent improvement across various metrics and benchmarks. For example, RMM shows more than 10% accuracy improvement over the baseline without memory management on the LongMemEval dataset.
Magnet: Multi-turn Tool-use Data Synthesis and Distillation via Graph Translation
Mar 10, 2025



Abstract:Large language models (LLMs) have exhibited the ability to effectively utilize external tools to address user queries. However, their performance may be limited in complex, multi-turn interactions involving users and multiple tools. To address this, we propose Magnet, a principled framework for synthesizing high-quality training trajectories to enhance the function calling capability of large language model agents in multi-turn conversations with humans. The framework is based on automatic and iterative translations from a function signature path to a sequence of queries and executable function calls. We model the complicated function interactions in multi-turn cases with graph and design novel node operations to build reliable signature paths. Motivated by context distillation, when guiding the generation of positive and negative trajectories using a teacher model, we provide reference function call sequences as positive hints in context and contrastive, incorrect function calls as negative hints. Experiments show that training with the positive trajectories with supervised fine-tuning and preference optimization against negative trajectories, our 14B model, Magnet-14B-mDPO, obtains 68.01 on BFCL-v3 and 73.30 on ToolQuery, surpassing the performance of the teacher model Gemini-1.5-pro-002 by a large margin in function calling.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge