Brandon Theodorou
InformGen: An AI Copilot for Accurate and Compliant Clinical Research Consent Document Generation
Apr 01, 2025Abstract:Leveraging large language models (LLMs) to generate high-stakes documents, such as informed consent forms (ICFs), remains a significant challenge due to the extreme need for regulatory compliance and factual accuracy. Here, we present InformGen, an LLM-driven copilot for accurate and compliant ICF drafting by optimized knowledge document parsing and content generation, with humans in the loop. We further construct a benchmark dataset comprising protocols and ICFs from 900 clinical trials. Experimental results demonstrate that InformGen achieves near 100% compliance with 18 core regulatory rules derived from FDA guidelines, outperforming a vanilla GPT-4o model by up to 30%. Additionally, a user study with five annotators shows that InformGen, when integrated with manual intervention, attains over 90% factual accuracy, significantly surpassing the vanilla GPT-4o model's 57%-82%. Crucially, InformGen ensures traceability by providing inline citations to source protocols, enabling easy verification and maintaining the highest standards of factual integrity.
ConSequence: Synthesizing Logically Constrained Sequences for Electronic Health Record Generation
Dec 20, 2023Abstract:Generative models can produce synthetic patient records for analytical tasks when real data is unavailable or limited. However, current methods struggle with adhering to domain-specific knowledge and removing invalid data. We present ConSequence, an effective approach to integrating domain knowledge into sequential generative neural network outputs. Our rule-based formulation includes temporal aggregation and antecedent evaluation modules, ensured by an efficient matrix multiplication formulation, to satisfy hard and soft logical constraints across time steps. Existing constraint methods often fail to guarantee constraint satisfaction, lack the ability to handle temporal constraints, and hinder the learning and computational efficiency of the model. In contrast, our approach efficiently handles all types of constraints with guaranteed logical coherence. We demonstrate ConSequence's effectiveness in generating electronic health records, outperforming competitors in achieving complete temporal and spatial constraint satisfaction without compromising runtime performance or generative quality. Specifically, ConSequence successfully prevents all rule violations while improving the model quality in reducing its test perplexity by 5% and incurring less than a 13% slowdown in generation speed compared to an unconstrained model.
TREEMENT: Interpretable Patient-Trial Matching via Personalized Dynamic Tree-Based Memory Network
Jul 19, 2023
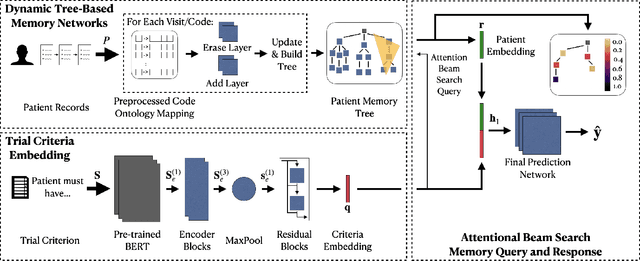
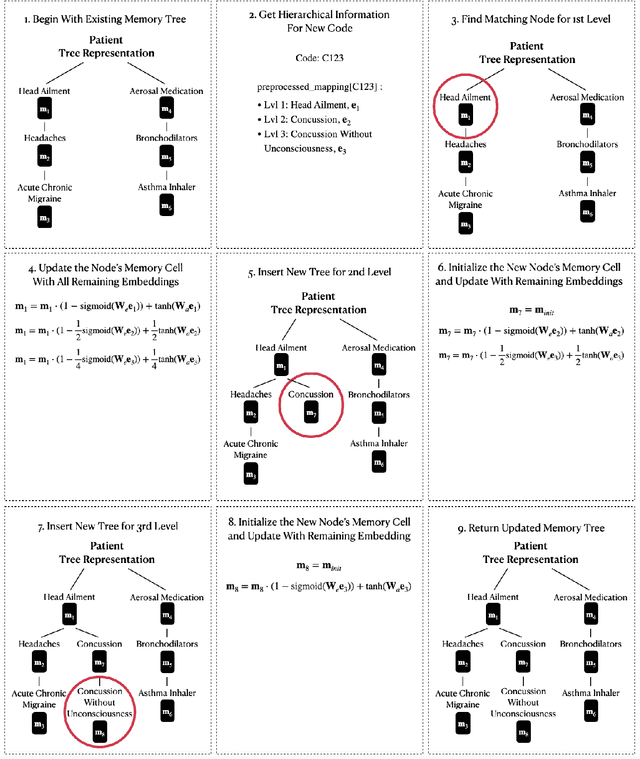
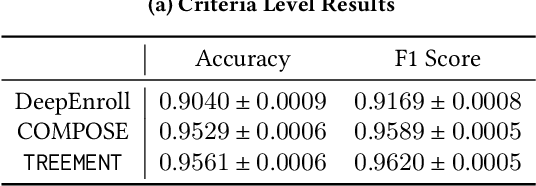
Abstract:Clinical trials are critical for drug development but often suffer from expensive and inefficient patient recruitment. In recent years, machine learning models have been proposed for speeding up patient recruitment via automatically matching patients with clinical trials based on longitudinal patient electronic health records (EHR) data and eligibility criteria of clinical trials. However, they either depend on trial-specific expert rules that cannot expand to other trials or perform matching at a very general level with a black-box model where the lack of interpretability makes the model results difficult to be adopted. To provide accurate and interpretable patient trial matching, we introduce a personalized dynamic tree-based memory network model named TREEMENT. It utilizes hierarchical clinical ontologies to expand the personalized patient representation learned from sequential EHR data, and then uses an attentional beam-search query learned from eligibility criteria embedding to offer a granular level of alignment for improved performance and interpretability. We evaluated TREEMENT against existing models on real-world datasets and demonstrated that TREEMENT outperforms the best baseline by 7% in terms of error reduction in criteria-level matching and achieves state-of-the-art results in its trial-level matching ability. Furthermore, we also show TREEMENT can offer good interpretability to make the model results easier for adoption.
PyTrial: A Comprehensive Platform for Artificial Intelligence for Drug Development
Jun 06, 2023



Abstract:Drug development is a complex process that aims to test the efficacy and safety of candidate drugs in the human body for regulatory approval via clinical trials. Recently, machine learning has emerged as a vital tool for drug development, offering new opportunities to improve the efficiency and success rates of the process. To facilitate the research and development of artificial intelligence (AI) for drug development, we developed a Python package, namely PyTrial, that implements various clinical trial tasks supported by AI algorithms. To be specific, PyTrial implements 6 essential drug development tasks, including patient outcome prediction, trial site selection, trial outcome prediction, patient-trial matching, trial similarity search, and synthetic data generation. In PyTrial, all tasks are defined by four steps: load data, model definition, model training, and model evaluation, which can be done with a couple of lines of code. In addition, the modular API design allows practitioners to extend the framework to new algorithms and tasks easily. PyTrial is featured for a unified API, detailed documentation, and interactive examples with preprocessed benchmark data for all implemented algorithms. This package can be installed through Python Package Index (PyPI) and is publicly available at https://github.com/RyanWangZf/PyTrial.
FRAMM: Fair Ranking with Missing Modalities for Clinical Trial Site Selection
May 30, 2023Abstract:Despite many efforts to address the disparities, the underrepresentation of gender, racial, and ethnic minorities in clinical trials remains a problem and undermines the efficacy of treatments on minorities. This paper focuses on the trial site selection task and proposes FRAMM, a deep reinforcement learning framework for fair trial site selection. We focus on addressing two real-world challenges that affect fair trial sites selection: the data modalities are often not complete for many potential trial sites, and the site selection needs to simultaneously optimize for both enrollment and diversity since the problem is necessarily a trade-off between the two with the only possible way to increase diversity post-selection being through limiting enrollment via caps. To address the missing data challenge, FRAMM has a modality encoder with a masked cross-attention mechanism for handling missing data, bypassing data imputation and the need for complete data in training. To handle the need for making efficient trade-offs, FRAMM uses deep reinforcement learning with a specifically designed reward function that simultaneously optimizes for both enrollment and fairness. We evaluate FRAMM using 4,392 real-world clinical trials ranging from 2016 to 2021 and show that FRAMM outperforms the leading baseline in enrollment-only settings while also achieving large gains in diversity. Specifically, it is able to produce a 9% improvement in diversity with similar enrollment levels over the leading baselines. That improved diversity is further manifested in achieving up to a 14% increase in Hispanic enrollment, 27% increase in Black enrollment, and 60% increase in Asian enrollment compared to selecting sites with an enrollment-only model.
Synthesize Extremely High-dimensional Longitudinal Electronic Health Records via Hierarchical Autoregressive Language Model
Apr 04, 2023Abstract:Synthetic electronic health records (EHRs) that are both realistic and preserve privacy can serve as an alternative to real EHRs for machine learning (ML) modeling and statistical analysis. However, generating high-fidelity and granular electronic health record (EHR) data in its original, highly-dimensional form poses challenges for existing methods due to the complexities inherent in high-dimensional data. In this paper, we propose Hierarchical Autoregressive Language mOdel (HALO) for generating longitudinal high-dimensional EHR, which preserve the statistical properties of real EHR and can be used to train accurate ML models without privacy concerns. Our HALO method, designed as a hierarchical autoregressive model, generates a probability density function of medical codes, clinical visits, and patient records, allowing for the generation of realistic EHR data in its original, unaggregated form without the need for variable selection or aggregation. Additionally, our model also produces high-quality continuous variables in a longitudinal and probabilistic manner. We conducted extensive experiments and demonstrate that HALO can generate high-fidelity EHR data with high-dimensional disease code probabilities (d > 10,000), disease co-occurrence probabilities within visits (d > 1,000,000), and conditional probabilities across consecutive visits (d > 5,000,000) and achieve above 0.9 R2 correlation in comparison to real EHR data. This performance then enables downstream ML models trained on its synthetic data to achieve comparable accuracy to models trained on real data (0.938 AUROC with HALO data vs. 0.943 with real data). Finally, using a combination of real and synthetic data enhances the accuracy of ML models beyond that achieved by using only real EHR data.
Clinical trial site matching with improved diversity using fair policy learning
Apr 13, 2022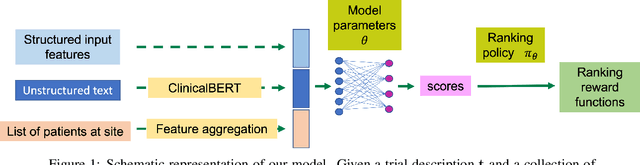
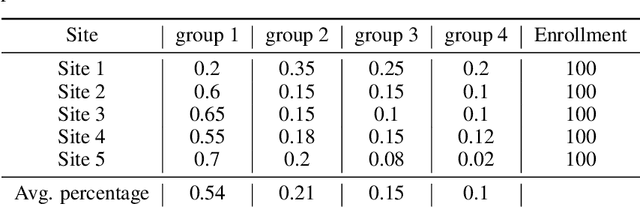
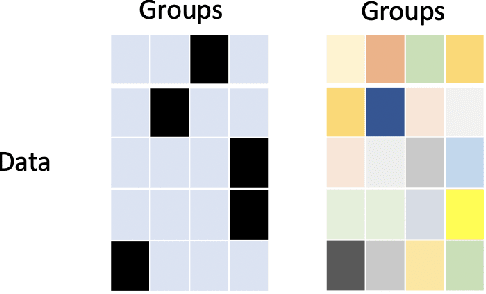
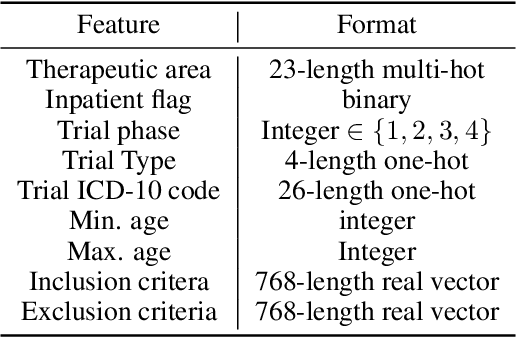
Abstract:The ongoing pandemic has highlighted the importance of reliable and efficient clinical trials in healthcare. Trial sites, where the trials are conducted, are chosen mainly based on feasibility in terms of medical expertise and access to a large group of patients. More recently, the issue of diversity and inclusion in clinical trials is gaining importance. Different patient groups may experience the effects of a medical drug/ treatment differently and hence need to be included in the clinical trials. These groups could be based on ethnicity, co-morbidities, age, or economic factors. Thus, designing a method for trial site selection that accounts for both feasibility and diversity is a crucial and urgent goal. In this paper, we formulate this problem as a ranking problem with fairness constraints. Using principles of fairness in machine learning, we learn a model that maps a clinical trial description to a ranked list of potential trial sites. Unlike existing fairness frameworks, the group membership of each trial site is non-binary: each trial site may have access to patients from multiple groups. We propose fairness criteria based on demographic parity to address such a multi-group membership scenario. We test our method on 480 real-world clinical trials and show that our model results in a list of potential trial sites that provides access to a diverse set of patients while also ensuing a high number of enrolled patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge