Lucas Glass
TrialBench: Multi-Modal Artificial Intelligence-Ready Clinical Trial Datasets
Jun 30, 2024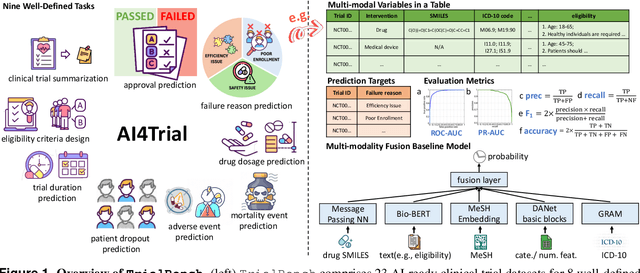

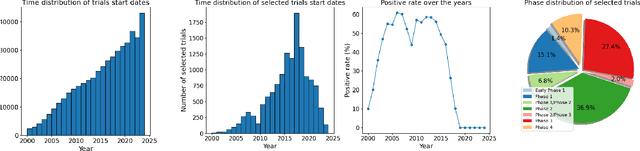
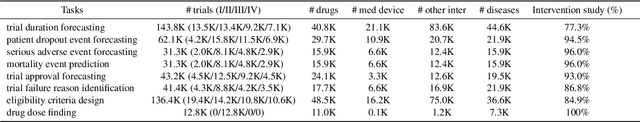
Abstract:Clinical trials are pivotal for developing new medical treatments, yet they typically pose some risks such as patient mortality, adverse events, and enrollment failure that waste immense efforts spanning over a decade. Applying artificial intelligence (AI) to forecast or simulate key events in clinical trials holds great potential for providing insights to guide trial designs. However, complex data collection and question definition requiring medical expertise and a deep understanding of trial designs have hindered the involvement of AI thus far. This paper tackles these challenges by presenting a comprehensive suite of meticulously curated AIready datasets covering multi-modal data (e.g., drug molecule, disease code, text, categorical/numerical features) and 8 crucial prediction challenges in clinical trial design, encompassing prediction of trial duration, patient dropout rate, serious adverse event, mortality rate, trial approval outcome, trial failure reason, drug dose finding, design of eligibility criteria. Furthermore, we provide basic validation methods for each task to ensure the datasets' usability and reliability. We anticipate that the availability of such open-access datasets will catalyze the development of advanced AI approaches for clinical trial design, ultimately advancing clinical trial research and accelerating medical solution development. The curated dataset, metrics, and basic models are publicly available at https://github.com/ML2Health/ML2ClinicalTrials/tree/main/AI4Trial.
Triple Simplex Matrix Completion for Expense Forecasting
Oct 23, 2023

Abstract:Forecasting project expenses is a crucial step for businesses to avoid budget overruns and project failures. Traditionally, this has been done by financial analysts or data science techniques such as time-series analysis. However, these approaches can be uncertain and produce results that differ from the planned budget, especially at the start of a project with limited data points. This paper proposes a constrained non-negative matrix completion model that predicts expenses by learning the likelihood of the project correlating with certain expense patterns in the latent space. The model is constrained on three probability simplexes, two of which are on the factor matrices and the third on the missing entries. Additionally, the predicted expense values are guaranteed to meet the budget constraint without the need of post-processing. An inexact alternating optimization algorithm is developed to solve the associated optimization problem and is proven to converge to a stationary point. Results from two real datasets demonstrate the effectiveness of the proposed method in comparison to state-of-the-art algorithms.
FRAMM: Fair Ranking with Missing Modalities for Clinical Trial Site Selection
May 30, 2023Abstract:Despite many efforts to address the disparities, the underrepresentation of gender, racial, and ethnic minorities in clinical trials remains a problem and undermines the efficacy of treatments on minorities. This paper focuses on the trial site selection task and proposes FRAMM, a deep reinforcement learning framework for fair trial site selection. We focus on addressing two real-world challenges that affect fair trial sites selection: the data modalities are often not complete for many potential trial sites, and the site selection needs to simultaneously optimize for both enrollment and diversity since the problem is necessarily a trade-off between the two with the only possible way to increase diversity post-selection being through limiting enrollment via caps. To address the missing data challenge, FRAMM has a modality encoder with a masked cross-attention mechanism for handling missing data, bypassing data imputation and the need for complete data in training. To handle the need for making efficient trade-offs, FRAMM uses deep reinforcement learning with a specifically designed reward function that simultaneously optimizes for both enrollment and fairness. We evaluate FRAMM using 4,392 real-world clinical trials ranging from 2016 to 2021 and show that FRAMM outperforms the leading baseline in enrollment-only settings while also achieving large gains in diversity. Specifically, it is able to produce a 9% improvement in diversity with similar enrollment levels over the leading baselines. That improved diversity is further manifested in achieving up to a 14% increase in Hispanic enrollment, 27% increase in Black enrollment, and 60% increase in Asian enrollment compared to selecting sites with an enrollment-only model.
DeCom: Deep Coupled-Factorization Machine for Post COVID-19 Respiratory Syncytial Virus Prediction with Nonpharmaceutical Interventions Awareness
May 02, 2023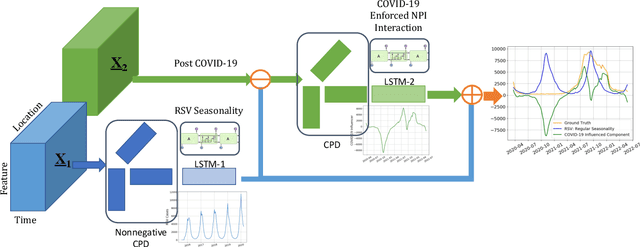
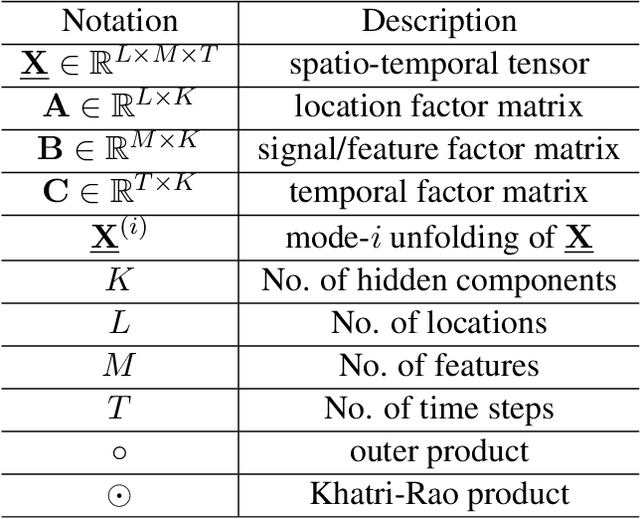
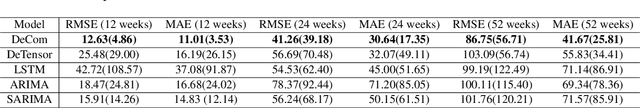
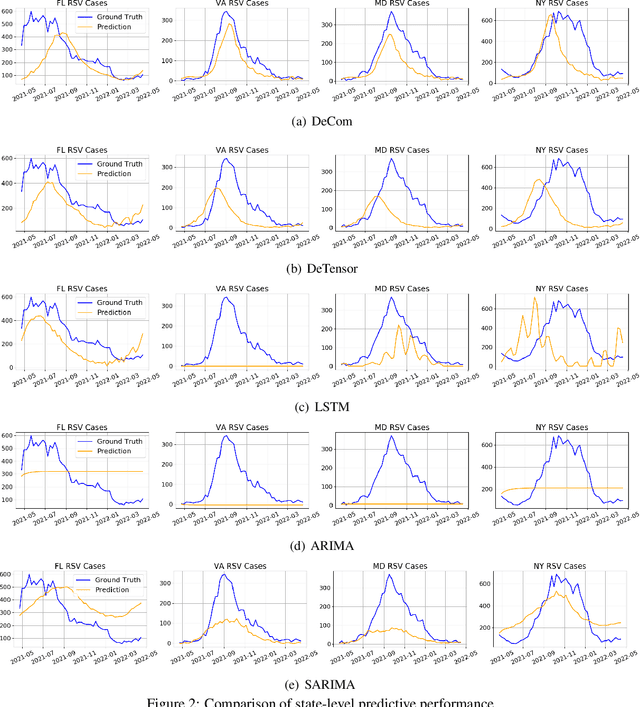
Abstract:Respiratory syncytial virus (RSV) is one of the most dangerous respiratory diseases for infants and young children. Due to the nonpharmaceutical intervention (NPI) imposed in the COVID-19 outbreak, the seasonal transmission pattern of RSV has been discontinued in 2020 and then shifted months ahead in 2021 in the northern hemisphere. It is critical to understand how COVID-19 impacts RSV and build predictive algorithms to forecast the timing and intensity of RSV reemergence in post-COVID-19 seasons. In this paper, we propose a deep coupled tensor factorization machine, dubbed as DeCom, for post COVID-19 RSV prediction. DeCom leverages tensor factorization and residual modeling. It enables us to learn the disrupted RSV transmission reliably under COVID-19 by taking both the regular seasonal RSV transmission pattern and the NPI into consideration. Experimental results on a real RSV dataset show that DeCom is more accurate than the state-of-the-art RSV prediction algorithms and achieves up to 46% lower root mean square error and 49% lower mean absolute error for country-level prediction compared to the baselines.
Clinical trial site matching with improved diversity using fair policy learning
Apr 13, 2022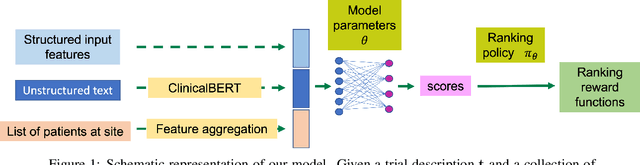
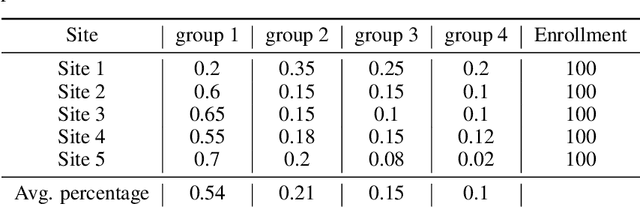
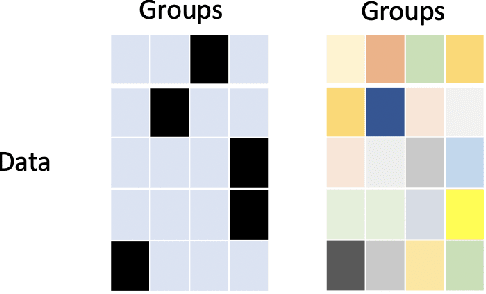
Abstract:The ongoing pandemic has highlighted the importance of reliable and efficient clinical trials in healthcare. Trial sites, where the trials are conducted, are chosen mainly based on feasibility in terms of medical expertise and access to a large group of patients. More recently, the issue of diversity and inclusion in clinical trials is gaining importance. Different patient groups may experience the effects of a medical drug/ treatment differently and hence need to be included in the clinical trials. These groups could be based on ethnicity, co-morbidities, age, or economic factors. Thus, designing a method for trial site selection that accounts for both feasibility and diversity is a crucial and urgent goal. In this paper, we formulate this problem as a ranking problem with fairness constraints. Using principles of fairness in machine learning, we learn a model that maps a clinical trial description to a ranked list of potential trial sites. Unlike existing fairness frameworks, the group membership of each trial site is non-binary: each trial site may have access to patients from multiple groups. We propose fairness criteria based on demographic parity to address such a multi-group membership scenario. We test our method on 480 real-world clinical trials and show that our model results in a list of potential trial sites that provides access to a diverse set of patients while also ensuing a high number of enrolled patients.
JULIA: Joint Multi-linear and Nonlinear Identification for Tensor Completion
Jan 31, 2022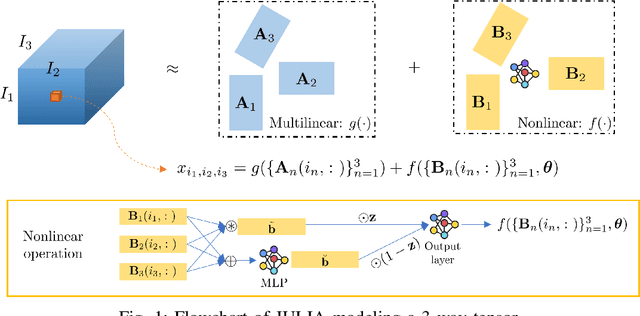
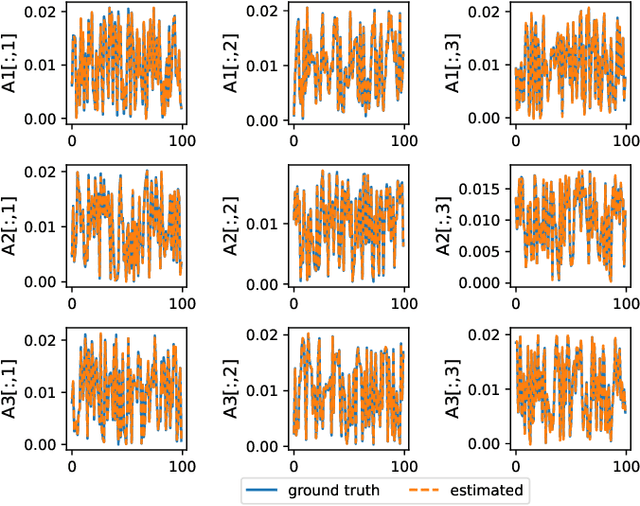
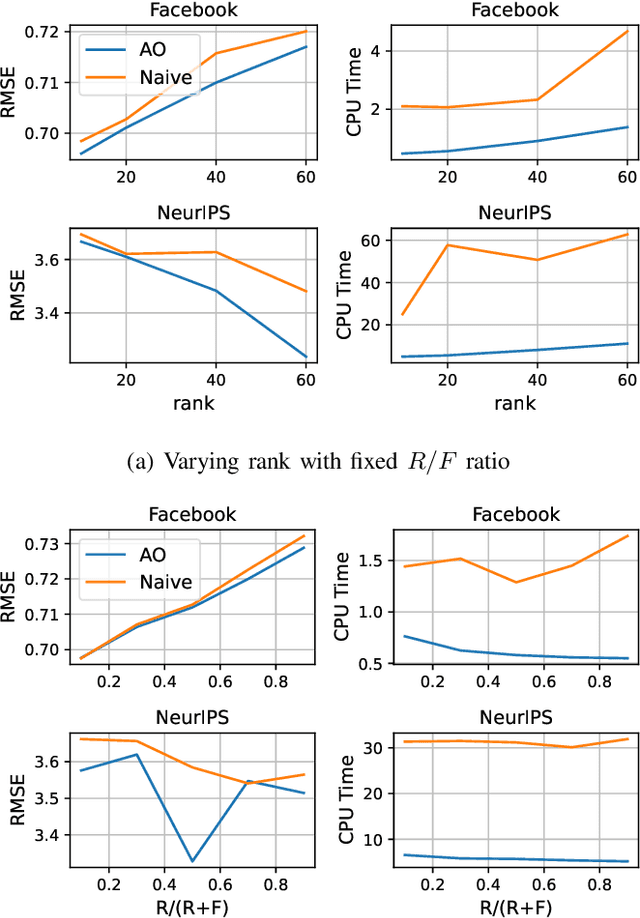
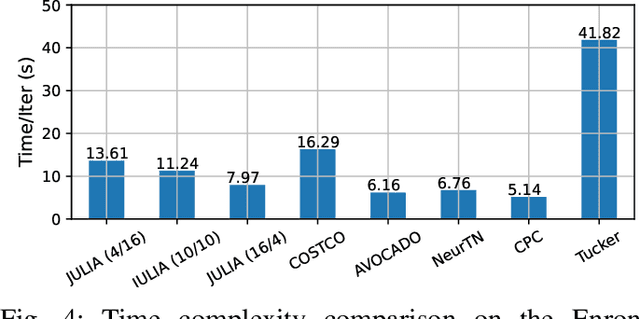
Abstract:Tensor completion aims at imputing missing entries from a partially observed tensor. Existing tensor completion methods often assume either multi-linear or nonlinear relationships between latent components. However, real-world tensors have much more complex patterns where both multi-linear and nonlinear relationships may coexist. In such cases, the existing methods are insufficient to describe the data structure. This paper proposes a Joint mUlti-linear and nonLinear IdentificAtion (JULIA) framework for large-scale tensor completion. JULIA unifies the multi-linear and nonlinear tensor completion models with several advantages over the existing methods: 1) Flexible model selection, i.e., it fits a tensor by assigning its values as a combination of multi-linear and nonlinear components; 2) Compatible with existing nonlinear tensor completion methods; 3) Efficient training based on a well-designed alternating optimization approach. Experiments on six real large-scale tensors demonstrate that JULIA outperforms many existing tensor completion algorithms. Furthermore, JULIA can improve the performance of a class of nonlinear tensor completion methods. The results show that in some large-scale tensor completion scenarios, baseline methods with JULIA are able to obtain up to 55% lower root mean-squared-error and save 67% computational complexity.
Multi-version Tensor Completion for Time-delayed Spatio-temporal Data
May 11, 2021



Abstract:Real-world spatio-temporal data is often incomplete or inaccurate due to various data loading delays. For example, a location-disease-time tensor of case counts can have multiple delayed updates of recent temporal slices for some locations or diseases. Recovering such missing or noisy (under-reported) elements of the input tensor can be viewed as a generalized tensor completion problem. Existing tensor completion methods usually assume that i) missing elements are randomly distributed and ii) noise for each tensor element is i.i.d. zero-mean. Both assumptions can be violated for spatio-temporal tensor data. We often observe multiple versions of the input tensor with different under-reporting noise levels. The amount of noise can be time- or location-dependent as more updates are progressively introduced to the tensor. We model such dynamic data as a multi-version tensor with an extra tensor mode capturing the data updates. We propose a low-rank tensor model to predict the updates over time. We demonstrate that our method can accurately predict the ground-truth values of many real-world tensors. We obtain up to 27.2% lower root mean-squared-error compared to the best baseline method. Finally, we extend our method to track the tensor data over time, leading to significant computational savings.
Change Matters: Medication Change Prediction with Recurrent Residual Networks
May 05, 2021
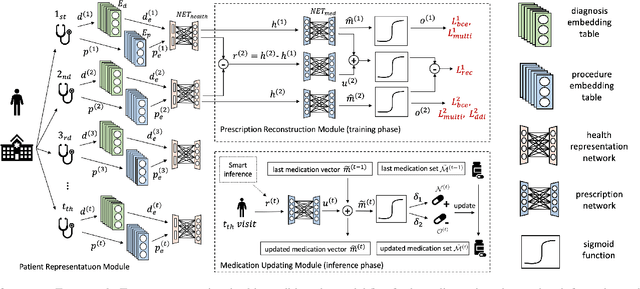

Abstract:Deep learning is revolutionizing predictive healthcare, including recommending medications to patients with complex health conditions. Existing approaches focus on predicting all medications for the current visit, which often overlaps with medications from previous visits. A more clinically relevant task is to identify medication changes. In this paper, we propose a new recurrent residual network, named MICRON, for medication change prediction. MICRON takes the changes in patient health records as input and learns to update a hidden medication vector and the medication set recurrently with a reconstruction design. The medication vector is like the memory cell that encodes longitudinal information of medications. Unlike traditional methods that require the entire patient history for prediction, MICRON has a residual-based inference that allows for sequential updating based only on new patient features (e.g., new diagnoses in the recent visit) more efficiently. We evaluated MICRON on real inpatient and outpatient datasets. MICRON achieves 3.5% and 7.8% relative improvements over the best baseline in F1 score, respectively. MICRON also requires fewer parameters, which significantly reduces the training time to 38.3s per epoch with 1.5x speed-up.
SafeDrug: Dual Molecular Graph Encoders for Safe Drug Recommendations
May 05, 2021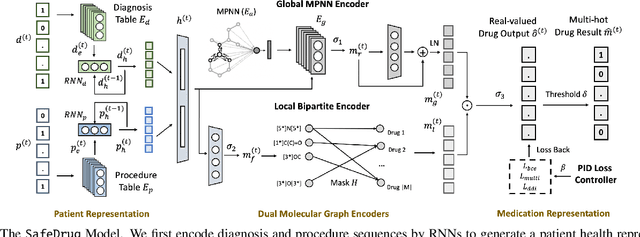
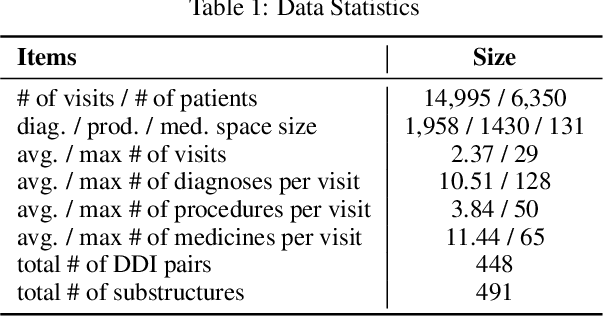

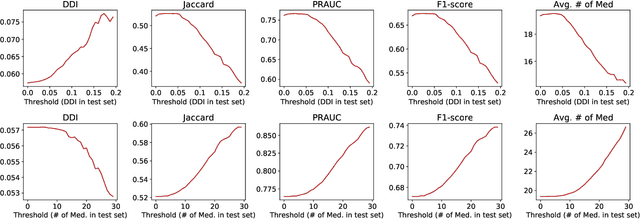
Abstract:Medication recommendation is an essential task of AI for healthcare. Existing works focused on recommending drug combinations for patients with complex health conditions solely based on their electronic health records. Thus, they have the following limitations: (1) some important data such as drug molecule structures have not been utilized in the recommendation process. (2) drug-drug interactions (DDI) are modeled implicitly, which can lead to sub-optimal results. To address these limitations, we propose a DDI-controllable drug recommendation model named SafeDrug to leverage drugs' molecule structures and model DDIs explicitly. SafeDrug is equipped with a global message passing neural network (MPNN) module and a local bipartite learning module to fully encode the connectivity and functionality of drug molecules. SafeDrug also has a controllable loss function to control DDI levels in the recommended drug combinations effectively. On a benchmark dataset, our SafeDrug is relatively shown to reduce DDI by 19.43% and improves 2.88% on Jaccard similarity between recommended and actually prescribed drug combinations over previous approaches. Moreover, SafeDrug also requires much fewer parameters than previous deep learning-based approaches, leading to faster training by about 14% and around 2x speed-up in inference.
SCRIB: Set-classifier with Class-specific Risk Bounds for Blackbox Models
Mar 05, 2021
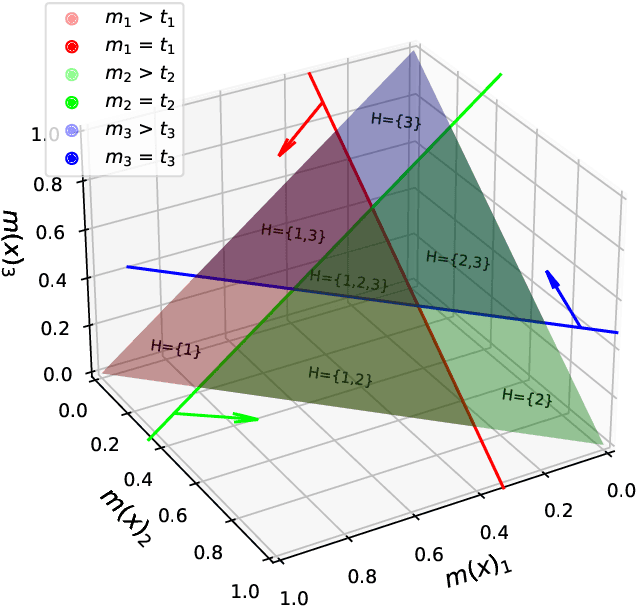
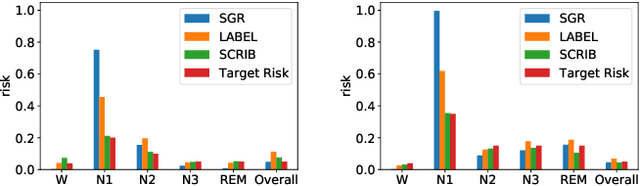

Abstract:Despite deep learning (DL) success in classification problems, DL classifiers do not provide a sound mechanism to decide when to refrain from predicting. Recent works tried to control the overall prediction risk with classification with rejection options. However, existing works overlook the different significance of different classes. We introduce Set-classifier with Class-specific RIsk Bounds (SCRIB) to tackle this problem, assigning multiple labels to each example. Given the output of a black-box model on the validation set, SCRIB constructs a set-classifier that controls the class-specific prediction risks with a theoretical guarantee. The key idea is to reject when the set classifier returns more than one label. We validated SCRIB on several medical applications, including sleep staging on electroencephalogram (EEG) data, X-ray COVID image classification, and atrial fibrillation detection based on electrocardiogram (ECG) data. SCRIB obtained desirable class-specific risks, which are 35\%-88\% closer to the target risks than baseline methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge