Miao Lin
RPP: A Certified Poisoned-Sample Detection Framework for Backdoor Attacks under Dataset Imbalance
Jan 30, 2026Abstract:Deep neural networks are highly susceptible to backdoor attacks, yet most defense methods to date rely on balanced data, overlooking the pervasive class imbalance in real-world scenarios that can amplify backdoor threats. This paper presents the first in-depth investigation of how the dataset imbalance amplifies backdoor vulnerability, showing that (i) the imbalance induces a majority-class bias that increases susceptibility and (ii) conventional defenses degrade significantly as the imbalance grows. To address this, we propose Randomized Probability Perturbation (RPP), a certified poisoned-sample detection framework that operates in a black-box setting using only model output probabilities. For any inspected sample, RPP determines whether the input has been backdoor-manipulated, while offering provable within-domain detectability guarantees and a probabilistic upper bound on the false positive rate. Extensive experiments on five benchmarks (MNIST, SVHN, CIFAR-10, TinyImageNet and ImageNet10) covering 10 backdoor attacks and 12 baseline defenses show that RPP achieves significantly higher detection accuracy than state-of-the-art defenses, particularly under dataset imbalance. RPP establishes a theoretical and practical foundation for defending against backdoor attacks in real-world environments with imbalanced data.
Multi-Modal CLIP-Informed Protein Editing
Jul 27, 2024Abstract:Proteins govern most biological functions essential for life, but achieving controllable protein discovery and optimization remains challenging. Recently, machine learning-assisted protein editing (MLPE) has shown promise in accelerating optimization cycles and reducing experimental workloads. However, current methods struggle with the vast combinatorial space of potential protein edits and cannot explicitly conduct protein editing using biotext instructions, limiting their interactivity with human feedback. To fill these gaps, we propose a novel method called ProtET for efficient CLIP-informed protein editing through multi-modality learning. Our approach comprises two stages: in the pretraining stage, contrastive learning aligns protein-biotext representations encoded by two large language models (LLMs), respectively. Subsequently, during the protein editing stage, the fused features from editing instruction texts and original protein sequences serve as the final editing condition for generating target protein sequences. Comprehensive experiments demonstrated the superiority of ProtET in editing proteins to enhance human-expected functionality across multiple attribute domains, including enzyme catalytic activity, protein stability and antibody specific binding ability. And ProtET improves the state-of-the-art results by a large margin, leading to significant stability improvements of 16.67% and 16.90%. This capability positions ProtET to advance real-world artificial protein editing, potentially addressing unmet academic, industrial, and clinical needs.
TrialBench: Multi-Modal Artificial Intelligence-Ready Clinical Trial Datasets
Jun 30, 2024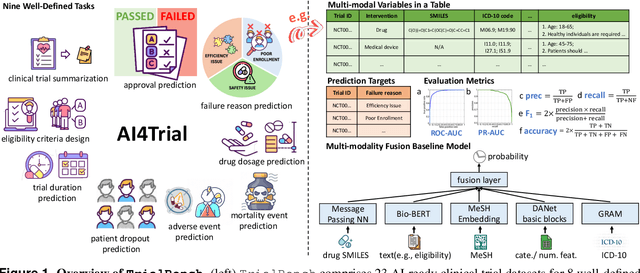

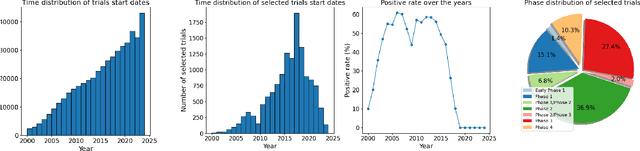
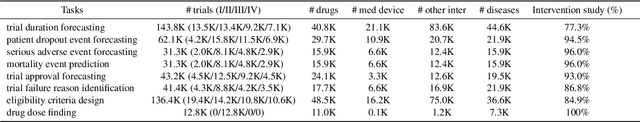
Abstract:Clinical trials are pivotal for developing new medical treatments, yet they typically pose some risks such as patient mortality, adverse events, and enrollment failure that waste immense efforts spanning over a decade. Applying artificial intelligence (AI) to forecast or simulate key events in clinical trials holds great potential for providing insights to guide trial designs. However, complex data collection and question definition requiring medical expertise and a deep understanding of trial designs have hindered the involvement of AI thus far. This paper tackles these challenges by presenting a comprehensive suite of meticulously curated AIready datasets covering multi-modal data (e.g., drug molecule, disease code, text, categorical/numerical features) and 8 crucial prediction challenges in clinical trial design, encompassing prediction of trial duration, patient dropout rate, serious adverse event, mortality rate, trial approval outcome, trial failure reason, drug dose finding, design of eligibility criteria. Furthermore, we provide basic validation methods for each task to ensure the datasets' usability and reliability. We anticipate that the availability of such open-access datasets will catalyze the development of advanced AI approaches for clinical trial design, ultimately advancing clinical trial research and accelerating medical solution development. The curated dataset, metrics, and basic models are publicly available at https://github.com/ML2Health/ML2ClinicalTrials/tree/main/AI4Trial.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge