Yaojun Hu
Real Doctor AI Research Centre, Zhejiang University, China
AnyECG: Foundational Models for Electrocardiogram Analysis
Nov 17, 2024



Abstract:Electrocardiogram (ECG), a non-invasive and affordable tool for cardiac monitoring, is highly sensitive in detecting acute heart attacks. However, due to the lengthy nature of ECG recordings, numerous machine learning methods have been developed for automated heart disease detection to reduce human workload. Despite these efforts, performance remains suboptimal. A key obstacle is the inherent complexity of ECG data, which includes heterogeneity (e.g., varying sampling rates), high levels of noise, demographic-related pattern shifts, and intricate rhythm-event associations. To overcome these challenges, this paper introduces AnyECG, a foundational model designed to extract robust representations from any real-world ECG data. Specifically, a tailored ECG Tokenizer encodes each fixed-duration ECG fragment into a token and, guided by proxy tasks, converts noisy, continuous ECG features into discrete, compact, and clinically meaningful local rhythm codes. These codes encapsulate basic morphological, frequency, and demographic information (e.g., sex), effectively mitigating signal noise. We further pre-train the AnyECG to learn rhythmic pattern associations across ECG tokens, enabling the capture of cardiac event semantics. By being jointly pre-trained on diverse ECG data sources, AnyECG is capable of generalizing across a wide range of downstream tasks where ECG signals are recorded from various devices and scenarios. Experimental results in anomaly detection, arrhythmia detection, corrupted lead generation, and ultra-long ECG signal analysis demonstrate that AnyECG learns common ECG knowledge from data and significantly outperforms cutting-edge methods in each respective task.
DrugCLIP: Contrastive Drug-Disease Interaction For Drug Repurposing
Jul 02, 2024

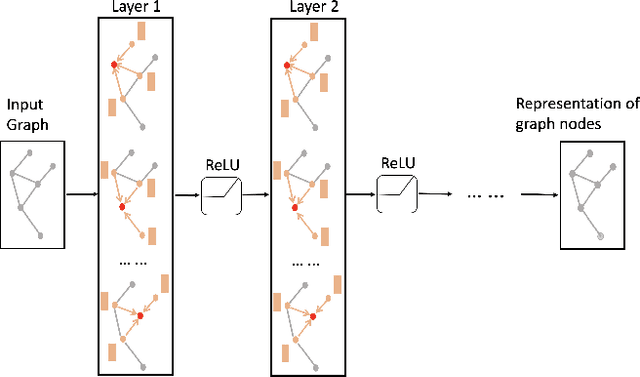

Abstract:Bringing a novel drug from the original idea to market typically requires more than ten years and billions of dollars. To alleviate the heavy burden, a natural idea is to reuse the approved drug to treat new diseases. The process is also known as drug repurposing or drug repositioning. Machine learning methods exhibited huge potential in automating drug repurposing. However, it still encounter some challenges, such as lack of labels and multimodal feature representation. To address these issues, we design DrugCLIP, a cutting-edge contrastive learning method, to learn drug and disease's interaction without negative labels. Additionally, we have curated a drug repurposing dataset based on real-world clinical trial records. Thorough empirical studies are conducted to validate the effectiveness of the proposed DrugCLIP method.
TrialBench: Multi-Modal Artificial Intelligence-Ready Clinical Trial Datasets
Jun 30, 2024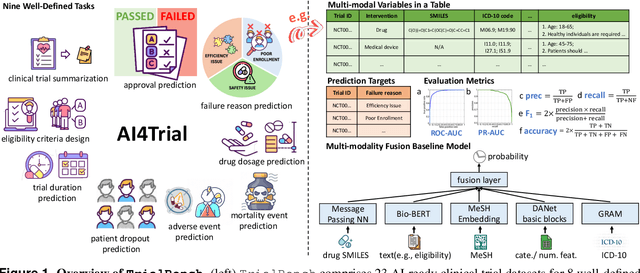

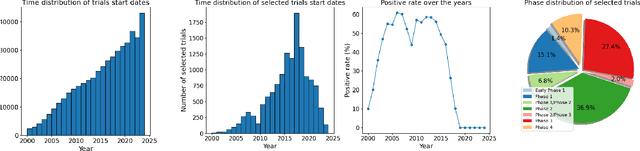
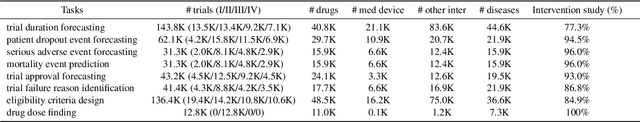
Abstract:Clinical trials are pivotal for developing new medical treatments, yet they typically pose some risks such as patient mortality, adverse events, and enrollment failure that waste immense efforts spanning over a decade. Applying artificial intelligence (AI) to forecast or simulate key events in clinical trials holds great potential for providing insights to guide trial designs. However, complex data collection and question definition requiring medical expertise and a deep understanding of trial designs have hindered the involvement of AI thus far. This paper tackles these challenges by presenting a comprehensive suite of meticulously curated AIready datasets covering multi-modal data (e.g., drug molecule, disease code, text, categorical/numerical features) and 8 crucial prediction challenges in clinical trial design, encompassing prediction of trial duration, patient dropout rate, serious adverse event, mortality rate, trial approval outcome, trial failure reason, drug dose finding, design of eligibility criteria. Furthermore, we provide basic validation methods for each task to ensure the datasets' usability and reliability. We anticipate that the availability of such open-access datasets will catalyze the development of advanced AI approaches for clinical trial design, ultimately advancing clinical trial research and accelerating medical solution development. The curated dataset, metrics, and basic models are publicly available at https://github.com/ML2Health/ML2ClinicalTrials/tree/main/AI4Trial.
Fair Evaluation of Federated Learning Algorithms for Automated Breast Density Classification: The Results of the 2022 ACR-NCI-NVIDIA Federated Learning Challenge
May 22, 2024Abstract:The correct interpretation of breast density is important in the assessment of breast cancer risk. AI has been shown capable of accurately predicting breast density, however, due to the differences in imaging characteristics across mammography systems, models built using data from one system do not generalize well to other systems. Though federated learning (FL) has emerged as a way to improve the generalizability of AI without the need to share data, the best way to preserve features from all training data during FL is an active area of research. To explore FL methodology, the breast density classification FL challenge was hosted in partnership with the American College of Radiology, Harvard Medical School's Mass General Brigham, University of Colorado, NVIDIA, and the National Institutes of Health National Cancer Institute. Challenge participants were able to submit docker containers capable of implementing FL on three simulated medical facilities, each containing a unique large mammography dataset. The breast density FL challenge ran from June 15 to September 5, 2022, attracting seven finalists from around the world. The winning FL submission reached a linear kappa score of 0.653 on the challenge test data and 0.413 on an external testing dataset, scoring comparably to a model trained on the same data in a central location.
* 16 pages, 9 figures
Personalized Heart Disease Detection via ECG Digital Twin Generation
Apr 17, 2024Abstract:Heart diseases rank among the leading causes of global mortality, demonstrating a crucial need for early diagnosis and intervention. Most traditional electrocardiogram (ECG) based automated diagnosis methods are trained at population level, neglecting the customization of personalized ECGs to enhance individual healthcare management. A potential solution to address this limitation is to employ digital twins to simulate symptoms of diseases in real patients. In this paper, we present an innovative prospective learning approach for personalized heart disease detection, which generates digital twins of healthy individuals' anomalous ECGs and enhances the model sensitivity to the personalized symptoms. In our approach, a vector quantized feature separator is proposed to locate and isolate the disease symptom and normal segments in ECG signals with ECG report guidance. Thus, the ECG digital twins can simulate specific heart diseases used to train a personalized heart disease detection model. Experiments demonstrate that our approach not only excels in generating high-fidelity ECG signals but also improves personalized heart disease detection. Moreover, our approach ensures robust privacy protection, safeguarding patient data in model development.
SERVAL: Synergy Learning between Vertical Models and LLMs towards Oracle-Level Zero-shot Medical Prediction
Mar 03, 2024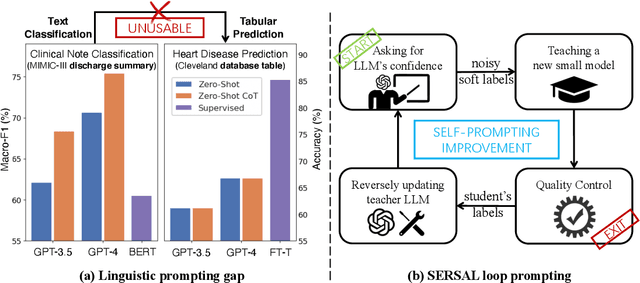

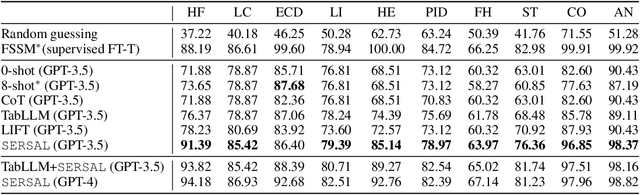
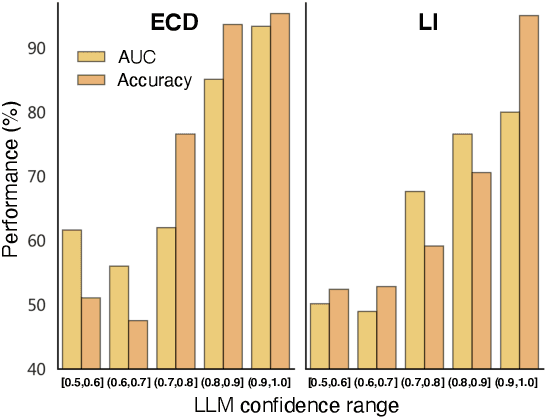
Abstract:Recent development of large language models (LLMs) has exhibited impressive zero-shot proficiency on generic and common sense questions. However, LLMs' application on domain-specific vertical questions still lags behind, primarily due to the humiliation problems and deficiencies in vertical knowledge. Furthermore, the vertical data annotation process often requires labor-intensive expert involvement, thereby presenting an additional challenge in enhancing the model's vertical capabilities. In this paper, we propose SERVAL, a synergy learning pipeline designed for unsupervised development of vertical capabilities in both LLMs and small models by mutual enhancement. Specifically, SERVAL utilizes the LLM's zero-shot outputs as annotations, leveraging its confidence to teach a robust vertical model from scratch. Reversely, the trained vertical model guides the LLM fine-tuning to enhance its zero-shot capability, progressively improving both models through an iterative process. In medical domain, known for complex vertical knowledge and costly annotations, comprehensive experiments show that, without access to any gold labels, SERVAL with the synergy learning of OpenAI GPT-3.5 and a simple model attains fully-supervised competitive performance across ten widely used medical datasets. These datasets represent vertically specialized medical diagnostic scenarios (e.g., diabetes, heart diseases, COVID-19), highlighting the potential of SERVAL in refining the vertical capabilities of LLMs and training vertical models from scratch, all achieved without the need for annotations.
Asymmetric Dual-Decoder U-Net for Joint Rain and Haze Removal
Jun 21, 2022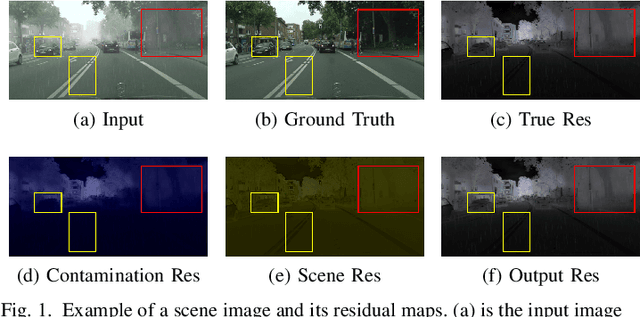


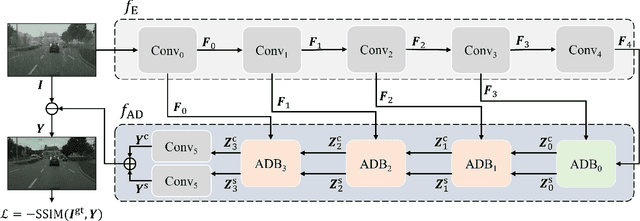
Abstract:This work studies the joint rain and haze removal problem. In real-life scenarios, rain and haze, two often co-occurring common weather phenomena, can greatly degrade the clarity and quality of the scene images, leading to a performance drop in the visual applications, such as autonomous driving. However, jointly removing the rain and haze in scene images is ill-posed and challenging, where the existence of haze and rain and the change of atmosphere light, can both degrade the scene information. Current methods focus on the contamination removal part, thus ignoring the restoration of the scene information affected by the change of atmospheric light. We propose a novel deep neural network, named Asymmetric Dual-decoder U-Net (ADU-Net), to address the aforementioned challenge. The ADU-Net produces both the contamination residual and the scene residual to efficiently remove the rain and haze while preserving the fidelity of the scene information. Extensive experiments show our work outperforms the existing state-of-the-art methods by a considerable margin in both synthetic data and real-world data benchmarks, including RainCityscapes, BID Rain, and SPA-Data. For instance, we improve the state-of-the-art PSNR value by 2.26/4.57 on the RainCityscapes/SPA-Data, respectively. Codes will be made available freely to the research community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge