Ken Chang
The Massachusetts General Hospital
SMMILE: An Expert-Driven Benchmark for Multimodal Medical In-Context Learning
Jun 26, 2025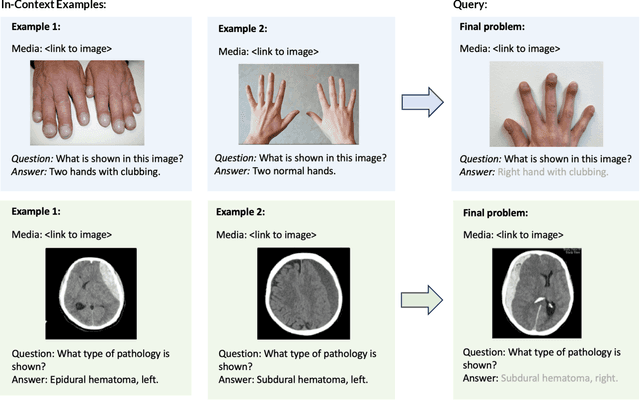



Abstract:Multimodal in-context learning (ICL) remains underexplored despite significant potential for domains such as medicine. Clinicians routinely encounter diverse, specialized tasks requiring adaptation from limited examples, such as drawing insights from a few relevant prior cases or considering a constrained set of differential diagnoses. While multimodal large language models (MLLMs) have shown advances in medical visual question answering (VQA), their ability to learn multimodal tasks from context is largely unknown. We introduce SMMILE, the first expert-driven multimodal ICL benchmark for medical tasks. Eleven medical experts curated problems, each including a multimodal query and multimodal in-context examples as task demonstrations. SMMILE encompasses 111 problems (517 question-image-answer triplets) covering 6 medical specialties and 13 imaging modalities. We further introduce SMMILE++, an augmented variant with 1038 permuted problems. A comprehensive evaluation of 15 MLLMs demonstrates that most models exhibit moderate to poor multimodal ICL ability in medical tasks. In open-ended evaluations, ICL contributes only 8% average improvement over zero-shot on SMMILE and 9.4% on SMMILE++. We observe a susceptibility for irrelevant in-context examples: even a single noisy or irrelevant example can degrade performance by up to 9.5%. Moreover, example ordering exhibits a recency bias, i.e., placing the most relevant example last can lead to substantial performance improvements by up to 71%. Our findings highlight critical limitations and biases in current MLLMs when learning multimodal medical tasks from context.
The 2024 Brain Tumor Segmentation (BraTS) Challenge: Glioma Segmentation on Post-treatment MRI
May 28, 2024
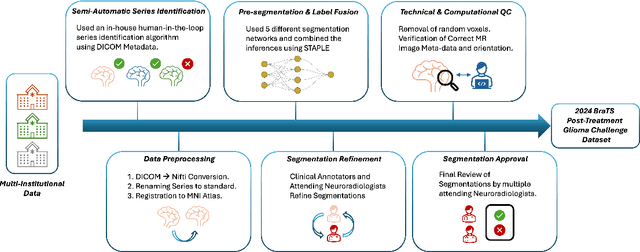
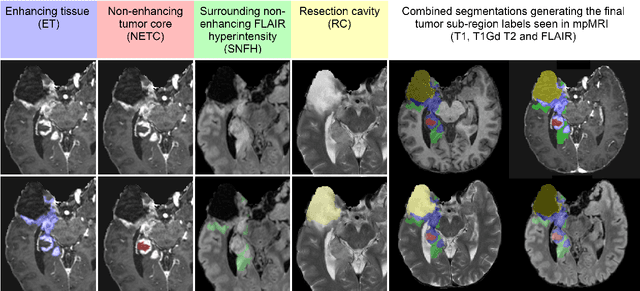
Abstract:Gliomas are the most common malignant primary brain tumors in adults and one of the deadliest types of cancer. There are many challenges in treatment and monitoring due to the genetic diversity and high intrinsic heterogeneity in appearance, shape, histology, and treatment response. Treatments include surgery, radiation, and systemic therapies, with magnetic resonance imaging (MRI) playing a key role in treatment planning and post-treatment longitudinal assessment. The 2024 Brain Tumor Segmentation (BraTS) challenge on post-treatment glioma MRI will provide a community standard and benchmark for state-of-the-art automated segmentation models based on the largest expert-annotated post-treatment glioma MRI dataset. Challenge competitors will develop automated segmentation models to predict four distinct tumor sub-regions consisting of enhancing tissue (ET), surrounding non-enhancing T2/fluid-attenuated inversion recovery (FLAIR) hyperintensity (SNFH), non-enhancing tumor core (NETC), and resection cavity (RC). Models will be evaluated on separate validation and test datasets using standardized performance metrics utilized across the BraTS 2024 cluster of challenges, including lesion-wise Dice Similarity Coefficient and Hausdorff Distance. Models developed during this challenge will advance the field of automated MRI segmentation and contribute to their integration into clinical practice, ultimately enhancing patient care.
Fair Evaluation of Federated Learning Algorithms for Automated Breast Density Classification: The Results of the 2022 ACR-NCI-NVIDIA Federated Learning Challenge
May 22, 2024Abstract:The correct interpretation of breast density is important in the assessment of breast cancer risk. AI has been shown capable of accurately predicting breast density, however, due to the differences in imaging characteristics across mammography systems, models built using data from one system do not generalize well to other systems. Though federated learning (FL) has emerged as a way to improve the generalizability of AI without the need to share data, the best way to preserve features from all training data during FL is an active area of research. To explore FL methodology, the breast density classification FL challenge was hosted in partnership with the American College of Radiology, Harvard Medical School's Mass General Brigham, University of Colorado, NVIDIA, and the National Institutes of Health National Cancer Institute. Challenge participants were able to submit docker containers capable of implementing FL on three simulated medical facilities, each containing a unique large mammography dataset. The breast density FL challenge ran from June 15 to September 5, 2022, attracting seven finalists from around the world. The winning FL submission reached a linear kappa score of 0.653 on the challenge test data and 0.413 on an external testing dataset, scoring comparably to a model trained on the same data in a central location.
* 16 pages, 9 figures
IMIL: Interactive Medical Image Learning Framework
Apr 17, 2024



Abstract:Data augmentations are widely used in training medical image deep learning models to increase the diversity and size of sparse datasets. However, commonly used augmentation techniques can result in loss of clinically relevant information from medical images, leading to incorrect predictions at inference time. We propose the Interactive Medical Image Learning (IMIL) framework, a novel approach for improving the training of medical image analysis algorithms that enables clinician-guided intermediate training data augmentations on misprediction outliers, focusing the algorithm on relevant visual information. To prevent the model from using irrelevant features during training, IMIL will 'blackout' clinician-designated irrelevant regions and replace the original images with the augmented samples. This ensures that for originally mispredicted samples, the algorithm subsequently attends only to relevant regions and correctly correlates them with the respective diagnosis. We validate the efficacy of IMIL using radiology residents and compare its performance to state-of-the-art data augmentations. A 4.2% improvement in accuracy over ResNet-50 was observed when using IMIL on only 4% of the training set. Our study demonstrates the utility of clinician-guided interactive training to achieve meaningful data augmentations for medical image analysis algorithms.
Towards More Efficient Data Valuation in Healthcare Federated Learning using Ensembling
Sep 12, 2022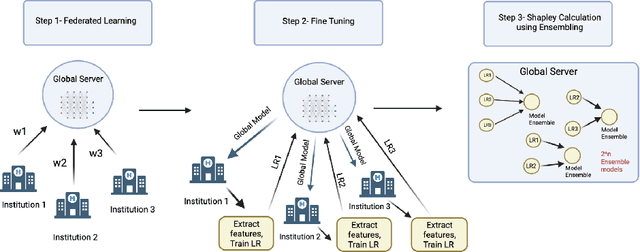
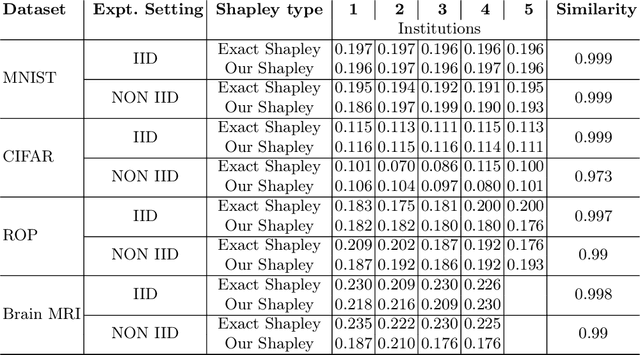

Abstract:Federated Learning (FL) wherein multiple institutions collaboratively train a machine learning model without sharing data is becoming popular. Participating institutions might not contribute equally, some contribute more data, some better quality data or some more diverse data. To fairly rank the contribution of different institutions, Shapley value (SV) has emerged as the method of choice. Exact SV computation is impossibly expensive, especially when there are hundreds of contributors. Existing SV computation techniques use approximations. However, in healthcare where the number of contributing institutions are likely not of a colossal scale, computing exact SVs is still exorbitantly expensive, but not impossible. For such settings, we propose an efficient SV computation technique called SaFE (Shapley Value for Federated Learning using Ensembling). We empirically show that SaFE computes values that are close to exact SVs, and that it performs better than current SV approximations. This is particularly relevant in medical imaging setting where widespread heterogeneity across institutions is rampant and fast accurate data valuation is required to determine the contribution of each participant in multi-institutional collaborative learning.
Three Applications of Conformal Prediction for Rating Breast Density in Mammography
Jun 23, 2022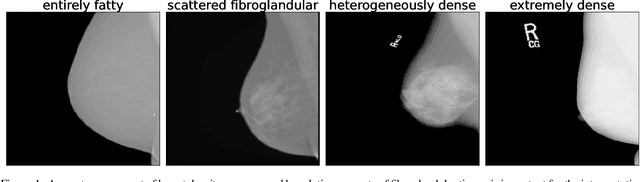
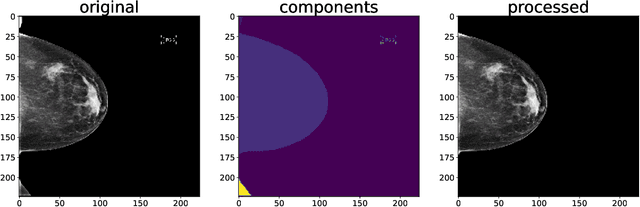
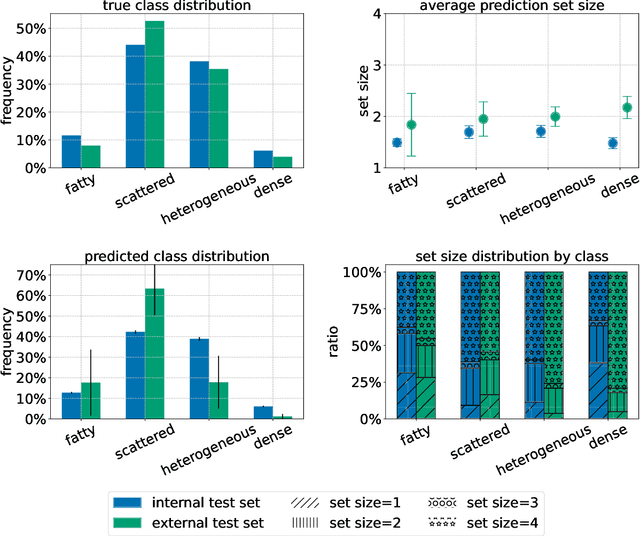
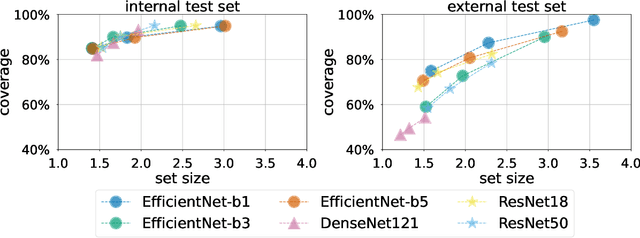
Abstract:Breast cancer is the most common cancers and early detection from mammography screening is crucial in improving patient outcomes. Assessing mammographic breast density is clinically important as the denser breasts have higher risk and are more likely to occlude tumors. Manual assessment by experts is both time-consuming and subject to inter-rater variability. As such, there has been increased interest in the development of deep learning methods for mammographic breast density assessment. Despite deep learning having demonstrated impressive performance in several prediction tasks for applications in mammography, clinical deployment of deep learning systems in still relatively rare; historically, mammography Computer-Aided Diagnoses (CAD) have over-promised and failed to deliver. This is in part due to the inability to intuitively quantify uncertainty of the algorithm for the clinician, which would greatly enhance usability. Conformal prediction is well suited to increase reliably and trust in deep learning tools but they lack realistic evaluations on medical datasets. In this paper, we present a detailed analysis of three possible applications of conformal prediction applied to medical imaging tasks: distribution shift characterization, prediction quality improvement, and subgroup fairness analysis. Our results show the potential of distribution-free uncertainty quantification techniques to enhance trust on AI algorithms and expedite their translation to usage.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
Decreasing Annotation Burden of Pairwise Comparisons with Human-in-the-Loop Sorting: Application in Medical Image Artifact Rating
Feb 10, 2022

Abstract:Ranking by pairwise comparisons has shown improved reliability over ordinal classification. However, as the annotations of pairwise comparisons scale quadratically, this becomes less practical when the dataset is large. We propose a method for reducing the number of pairwise comparisons required to rank by a quantitative metric, demonstrating the effectiveness of the approach in ranking medical images by image quality in this proof of concept study. Using the medical image annotation software that we developed, we actively subsample pairwise comparisons using a sorting algorithm with a human rater in the loop. We find that this method substantially reduces the number of comparisons required for a full ordinal ranking without compromising inter-rater reliability when compared to pairwise comparisons without sorting.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021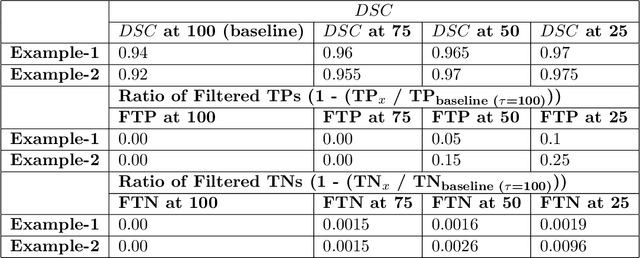
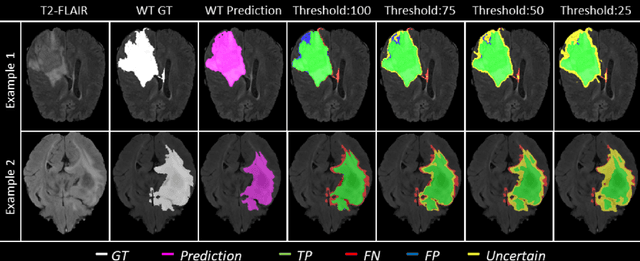
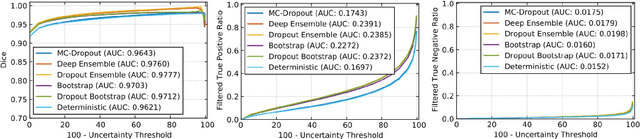
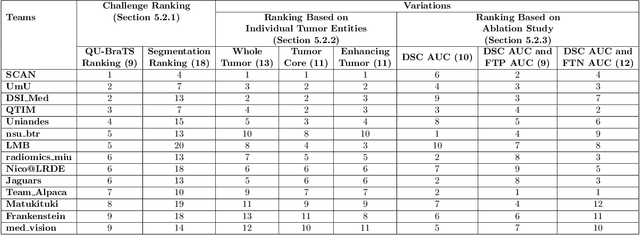
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Deploying clinical machine learning? Consider the following...
Sep 14, 2021
Abstract:Despite the intense attention and investment into clinical machine learning (CML) research, relatively few applications convert to clinical practice. While research is important in advancing the state-of-the-art, translation is equally important in bringing these technologies into a position to ultimately impact patient care and live up to extensive expectations surrounding AI in healthcare. To better characterize a holistic perspective among researchers and practitioners, we survey several participants with experience in developing CML for clinical deployment about their learned experiences. We collate these insights and identify several main categories of barriers and pitfalls in order to better design and develop clinical machine learning applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge