Katharina Hoebel
Improving the repeatability of deep learning models with Monte Carlo dropout
Feb 15, 2022



Abstract:The integration of artificial intelligence into clinical workflows requires reliable and robust models. Repeatability is a key attribute of model robustness. Repeatable models output predictions with low variation during independent tests carried out under similar conditions. During model development and evaluation, much attention is given to classification performance while model repeatability is rarely assessed, leading to the development of models that are unusable in clinical practice. In this work, we evaluate the repeatability of four model types (binary classification, multi-class classification, ordinal classification, and regression) on images that were acquired from the same patient during the same visit. We study the performance of binary, multi-class, ordinal, and regression models on four medical image classification tasks from public and private datasets: knee osteoarthritis, cervical cancer screening, breast density estimation, and retinopathy of prematurity. Repeatability is measured and compared on ResNet and DenseNet architectures. Moreover, we assess the impact of sampling Monte Carlo dropout predictions at test time on classification performance and repeatability. Leveraging Monte Carlo predictions significantly increased repeatability for all tasks on the binary, multi-class, and ordinal models leading to an average reduction of the 95\% limits of agreement by 16% points and of the disagreement rate by 7% points. The classification accuracy improved in most settings along with the repeatability. Our results suggest that beyond about 20 Monte Carlo iterations, there is no further gain in repeatability. In addition to the higher test-retest agreement, Monte Carlo predictions were better calibrated which leads to output probabilities reflecting more accurately the true likelihood of being correctly classified.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021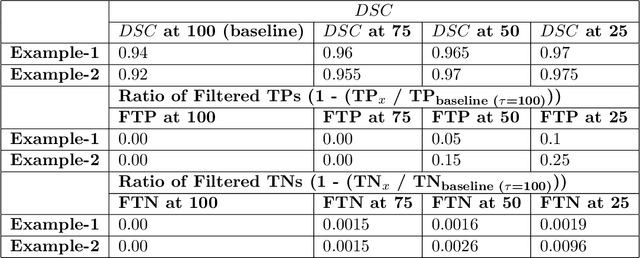
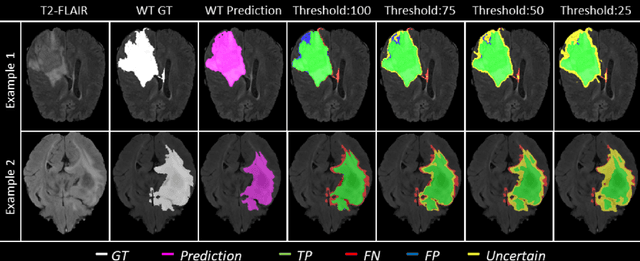
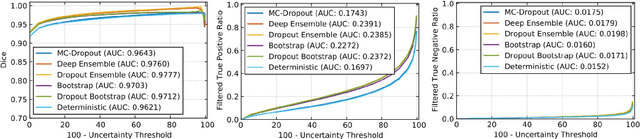
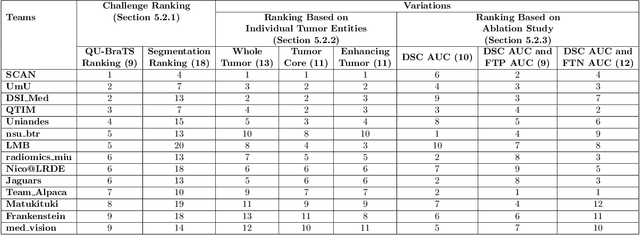
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Monte Carlo dropout increases model repeatability
Nov 12, 2021



Abstract:The integration of artificial intelligence into clinical workflows requires reliable and robust models. Among the main features of robustness is repeatability. Much attention is given to classification performance without assessing the model repeatability, leading to the development of models that turn out to be unusable in practice. In this work, we evaluate the repeatability of four model types on images from the same patient that were acquired during the same visit. We study the performance of binary, multi-class, ordinal, and regression models on three medical image analysis tasks: cervical cancer screening, breast density estimation, and retinopathy of prematurity classification. Moreover, we assess the impact of sampling Monte Carlo dropout predictions at test time on classification performance and repeatability. Leveraging Monte Carlo predictions significantly increased repeatability for all tasks on the binary, multi-class, and ordinal models leading to an average reduction of the 95% limits of agreement by 17% points.
Fair Conformal Predictors for Applications in Medical Imaging
Sep 09, 2021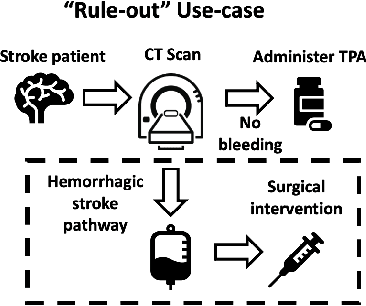
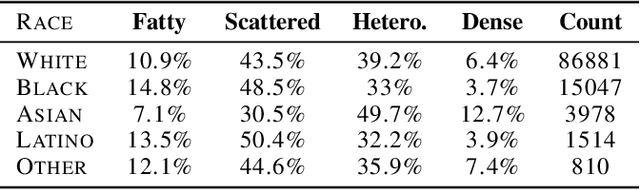
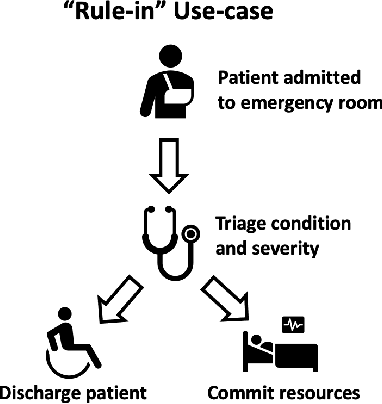
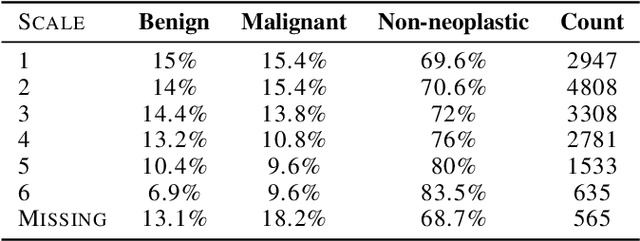
Abstract:Deep learning has the potential to augment many components of the clinical workflow, such as medical image interpretation. However, the translation of these black box algorithms into clinical practice has been marred by the relative lack of transparency compared to conventional machine learning methods, hindering in clinician trust in the systems for critical medical decision-making. Specifically, common deep learning approaches do not have intuitive ways of expressing uncertainty with respect to cases that might require further human review. Furthermore, the possibility of algorithmic bias has caused hesitancy regarding the use of developed algorithms in clinical settings. To these ends, we explore how conformal methods can complement deep learning models by providing both clinically intuitive way (by means of confidence prediction sets) of expressing model uncertainty as well as facilitating model transparency in clinical workflows. In this paper, we conduct a field survey with clinicians to assess clinical use-cases of conformal predictions. Next, we conduct experiments with a mammographic breast density and dermatology photography datasets to demonstrate the utility of conformal predictions in "rule-in" and "rule-out" disease scenarios. Further, we show that conformal predictors can be used to equalize coverage with respect to patient demographics such as race and skin tone. We find that a conformal predictions to be a promising framework with potential to increase clinical usability and transparency for better collaboration between deep learning algorithms and clinicians.
Evaluating subgroup disparity using epistemic uncertainty in mammography
Jul 15, 2021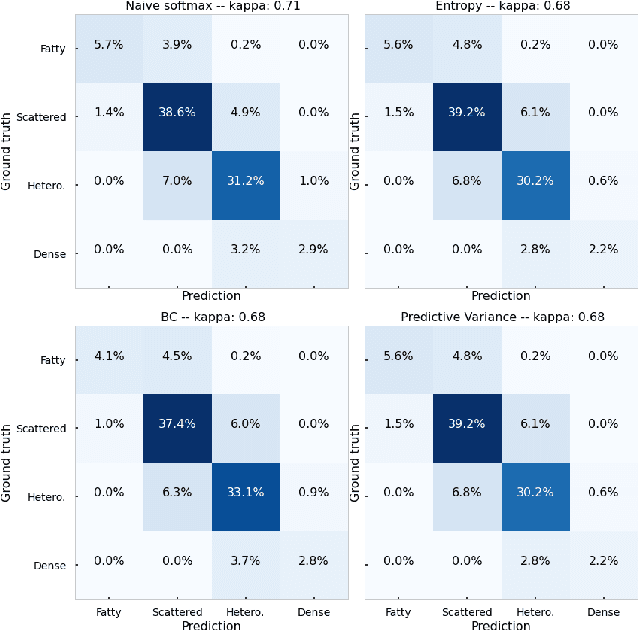
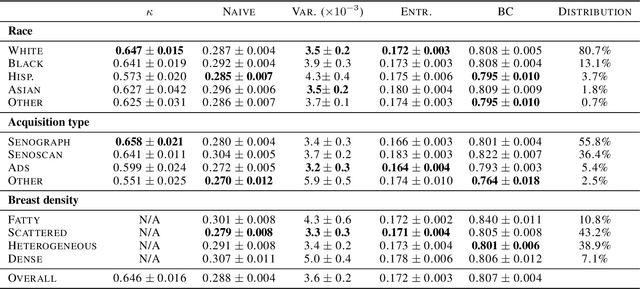
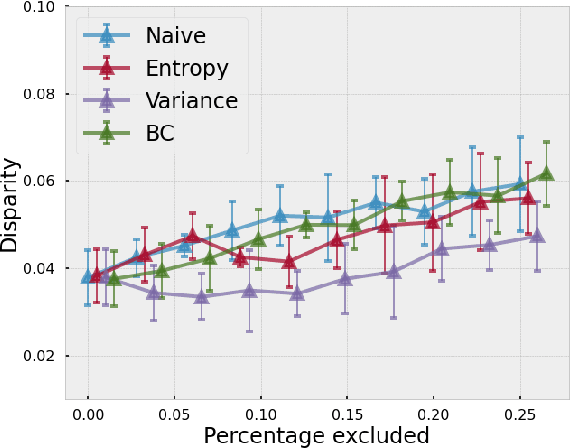
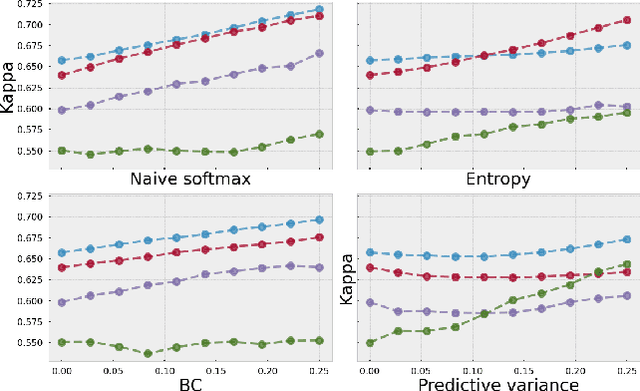
Abstract:As machine learning (ML) continue to be integrated into healthcare systems that affect clinical decision making, new strategies will need to be incorporated in order to effectively detect and evaluate subgroup disparities to ensure accountability and generalizability in clinical workflows. In this paper, we explore how epistemic uncertainty can be used to evaluate disparity in patient demographics (race) and data acquisition (scanner) subgroups for breast density assessment on a dataset of 108,190 mammograms collected from 33 clinical sites. Our results show that even if aggregate performance is comparable, the choice of uncertainty quantification metric can significantly the subgroup level. We hope this analysis can promote further work on how uncertainty can be leveraged to increase transparency of machine learning applications for clinical deployment.
Addressing catastrophic forgetting for medical domain expansion
Mar 24, 2021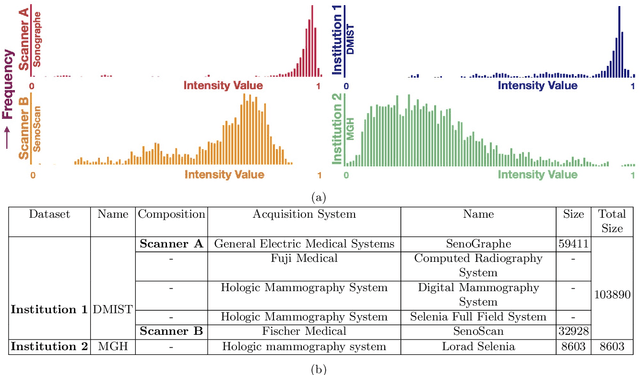
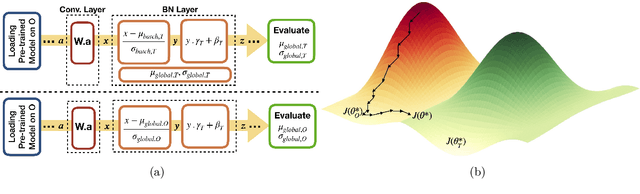
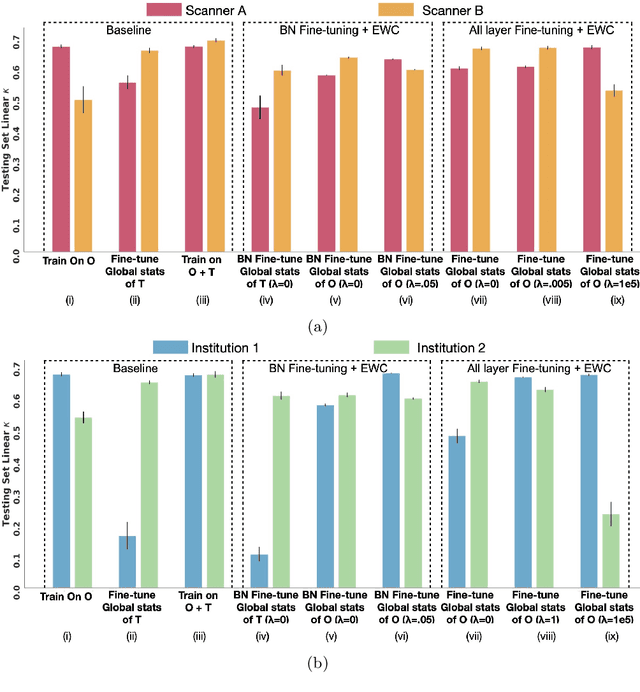
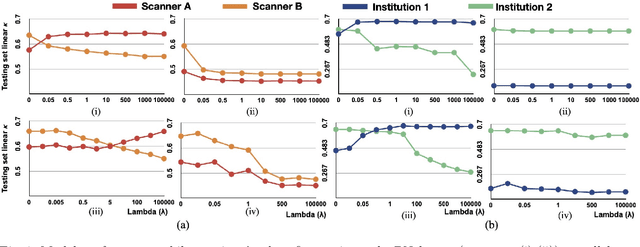
Abstract:Model brittleness is a key concern when deploying deep learning models in real-world medical settings. A model that has high performance at one institution may suffer a significant decline in performance when tested at other institutions. While pooling datasets from multiple institutions and retraining may provide a straightforward solution, it is often infeasible and may compromise patient privacy. An alternative approach is to fine-tune the model on subsequent institutions after training on the original institution. Notably, this approach degrades model performance at the original institution, a phenomenon known as catastrophic forgetting. In this paper, we develop an approach to address catastrophic forget-ting based on elastic weight consolidation combined with modulation of batch normalization statistics under two scenarios: first, for expanding the domain from one imaging system's data to another imaging system's, and second, for expanding the domain from a large multi-institutional dataset to another single institution dataset. We show that our approach outperforms several other state-of-the-art approaches and provide theoretical justification for the efficacy of batch normalization modulation. The results of this study are generally applicable to the deployment of any clinical deep learning model which requires domain expansion.
The unreasonable effectiveness of Batch-Norm statistics in addressing catastrophic forgetting across medical institutions
Nov 16, 2020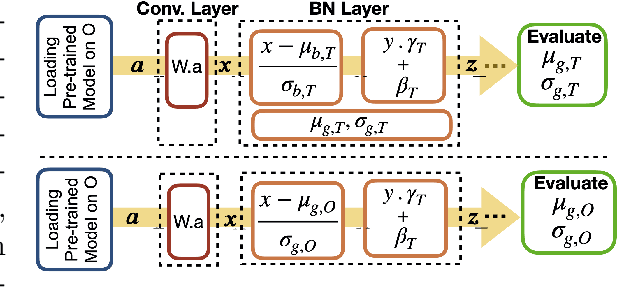
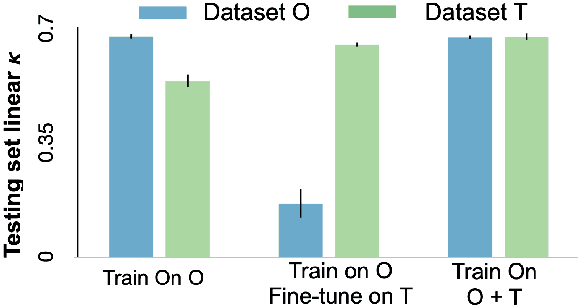
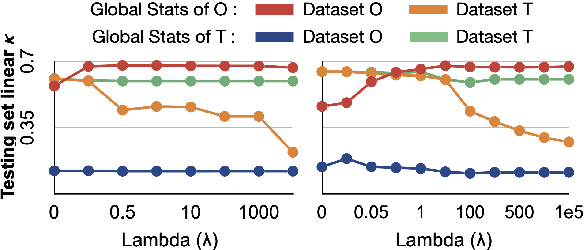
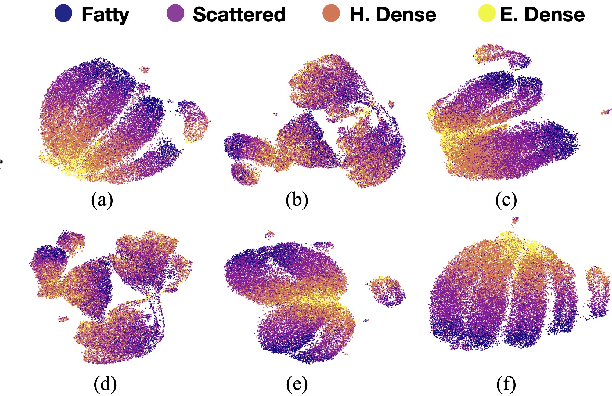
Abstract:Model brittleness is a primary concern when deploying deep learning models in medical settings owing to inter-institution variations, like patient demographics and intra-institution variation, such as multiple scanner types. While simply training on the combined datasets is fraught with data privacy limitations, fine-tuning the model on subsequent institutions after training it on the original institution results in a decrease in performance on the original dataset, a phenomenon called catastrophic forgetting. In this paper, we investigate trade-off between model refinement and retention of previously learned knowledge and subsequently address catastrophic forgetting for the assessment of mammographic breast density. More specifically, we propose a simple yet effective approach, adapting Elastic weight consolidation (EWC) using the global batch normalization (BN) statistics of the original dataset. The results of this study provide guidance for the deployment of clinical deep learning models where continuous learning is needed for domain expansion.
Assessing the (Un)Trustworthiness of Saliency Maps for Localizing Abnormalities in Medical Imaging
Aug 06, 2020Abstract:Saliency maps have become a widely used method to make deep learning models more interpretable by providing post-hoc explanations of classifiers through identification of the most pertinent areas of the input medical image. They are increasingly being used in medical imaging to provide clinically plausible explanations for the decisions the neural network makes. However, the utility and robustness of these visualization maps has not yet been rigorously examined in the context of medical imaging. We posit that trustworthiness in this context requires 1) localization utility, 2) sensitivity to model weight randomization, 3) repeatability, and 4) reproducibility. Using the localization information available in two large public radiology datasets, we quantify the performance of eight commonly used saliency map approaches for the above criteria using area under the precision-recall curves (AUPRC) and structural similarity index (SSIM), comparing their performance to various baseline measures. Using our framework to quantify the trustworthiness of saliency maps, we show that all eight saliency map techniques fail at least one of the criteria and are, in most cases, less trustworthy when compared to the baselines. We suggest that their usage in the high-risk domain of medical imaging warrants additional scrutiny and recommend that detection or segmentation models be used if localization is the desired output of the network. Additionally, to promote reproducibility of our findings, we provide the code we used for all tests performed in this work at this link: https://github.com/QTIM-Lab/Assessing-Saliency-Maps.
Give me (un)certainty -- An exploration of parameters that affect segmentation uncertainty
Nov 14, 2019

Abstract:Segmentation tasks in medical imaging are inherently ambiguous: the boundary of a target structure is oftentimes unclear due to image quality and biological factors. As such, predicted segmentations from deep learning algorithms are inherently ambiguous. Additionally, "ground truth" segmentations performed by human annotators are in fact weak labels that further increase the uncertainty of outputs of supervised models developed on these manual labels. To date, most deep learning segmentation studies utilize predicted segmentations without uncertainty quantification. In contrast, we explore the use of Monte Carlo dropout U-Nets for the segmentation with additional quantification of segmentation uncertainty. We assess the utility of three measures of uncertainty (Coefficient of Variation, Mean Pairwise Dice, and Mean Voxelwise Uncertainty) for the segmentation of a less ambiguous target structure (liver) and a more ambiguous one (liver tumors). Furthermore, we assess how the utility of these measures changes with different patch sizes and cost functions. Our results suggest that models trained using larger patches and the weighted categorical cross-entropy as cost function allow the extraction of more meaningful uncertainty measures compared to smaller patches and soft dice loss. Among the three uncertainty measures Mean Pairwise Dice shows the strongest correlation with segmentation quality. Our study serves as a proof-of-concept of how uncertainty measures can be used to assess the quality of a predicted segmentation, potentially serving to flag low quality segmentations from a given model for further human review.
DeepNeuro: an open-source deep learning toolbox for neuroimaging
Aug 14, 2018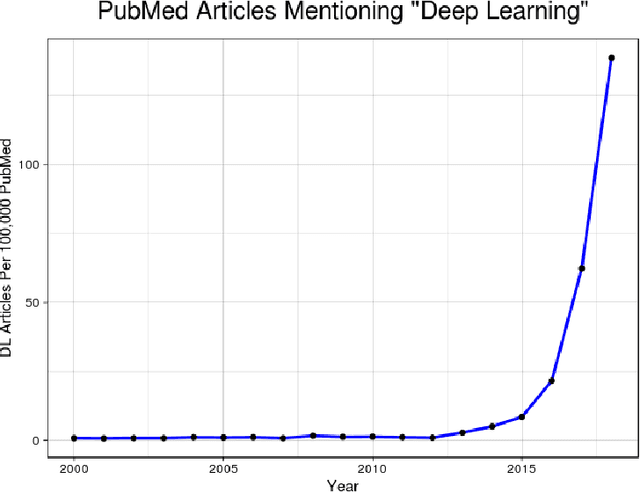
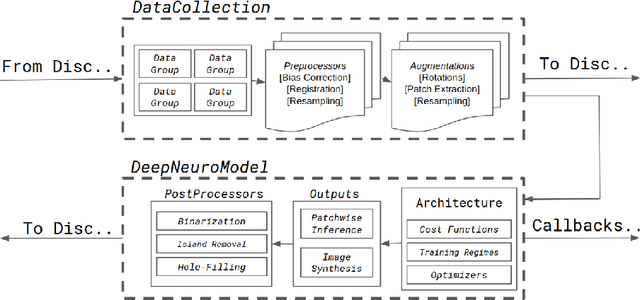
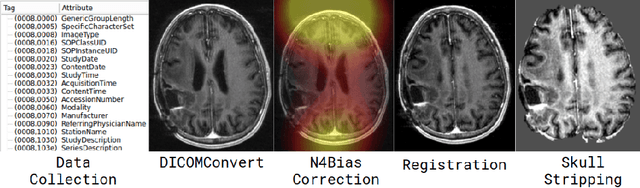
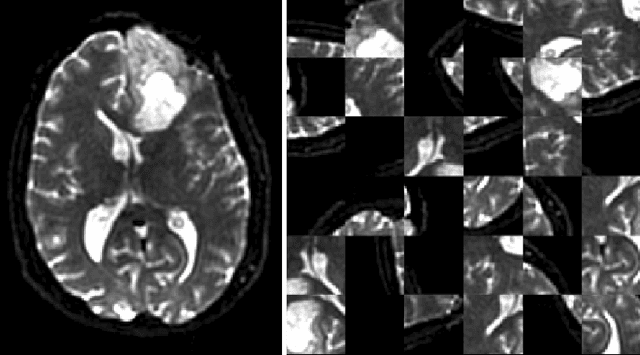
Abstract:Translating neural networks from theory to clinical practice has unique challenges, specifically in the field of neuroimaging. In this paper, we present DeepNeuro, a deep learning framework that is best-suited to putting deep learning algorithms for neuroimaging in practical usage with a minimum of friction. We show how this framework can be used to both design and train neural network architectures, as well as modify state-of-the-art architectures in a flexible and intuitive way. We display the pre- and postprocessing functions common in the medical imaging community that DeepNeuro offers to ensure consistent performance of networks across variable users, institutions, and scanners. And we show how pipelines created in DeepNeuro can be concisely packaged into shareable Docker containers and command-line interfaces using DeepNeuro's pipeline resources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge