Ashwin Vaswani
All Mistakes Are Not Equal: Comprehensive Hierarchy Aware Multi-label Predictions (CHAMP)
Jun 17, 2022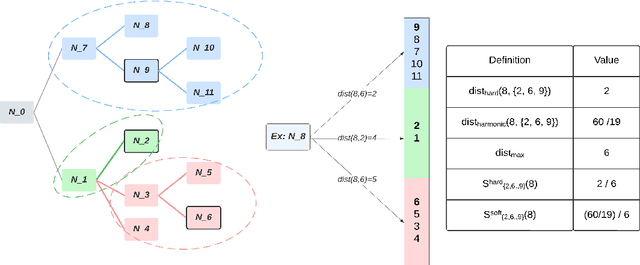
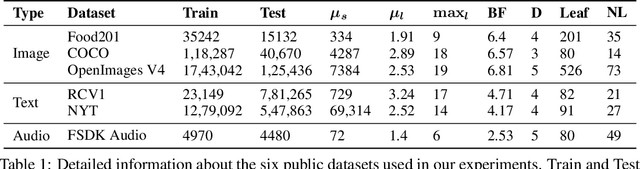
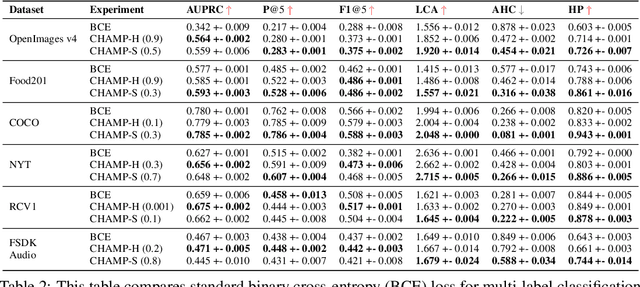
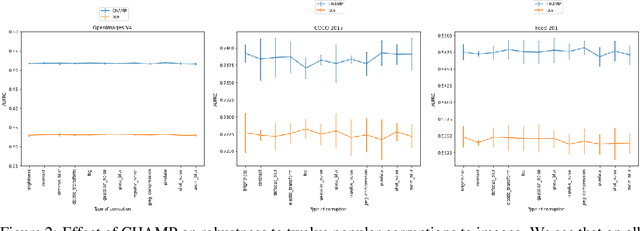
Abstract:This paper considers the problem of Hierarchical Multi-Label Classification (HMC), where (i) several labels can be present for each example, and (ii) labels are related via a domain-specific hierarchy tree. Guided by the intuition that all mistakes are not equal, we present Comprehensive Hierarchy Aware Multi-label Predictions (CHAMP), a framework that penalizes a misprediction depending on its severity as per the hierarchy tree. While there have been works that apply such an idea to single-label classification, to the best of our knowledge, there are limited such works for multilabel classification focusing on the severity of mistakes. The key reason is that there is no clear way of quantifying the severity of a misprediction a priori in the multilabel setting. In this work, we propose a simple but effective metric to quantify the severity of a mistake in HMC, naturally leading to CHAMP. Extensive experiments on six public HMC datasets across modalities (image, audio, and text) demonstrate that incorporating hierarchical information leads to substantial gains as CHAMP improves both AUPRC (2.6% median percentage improvement) and hierarchical metrics (2.85% median percentage improvement), over stand-alone hierarchical or multilabel classification methods. Compared to standard multilabel baselines, CHAMP provides improved AUPRC in both robustness (8.87% mean percentage improvement ) and less data regimes. Further, our method provides a framework to enhance existing multilabel classification algorithms with better mistakes (18.1% mean percentage increment).
Addressing catastrophic forgetting for medical domain expansion
Mar 24, 2021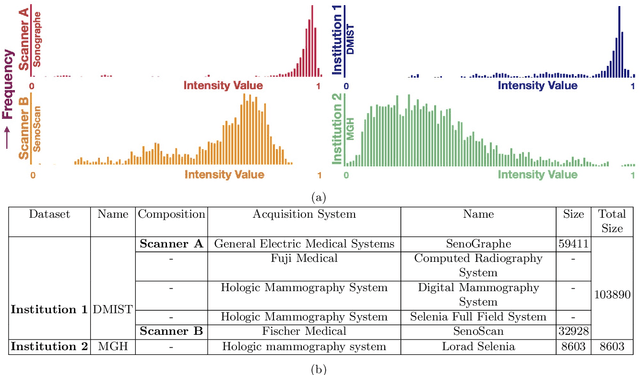
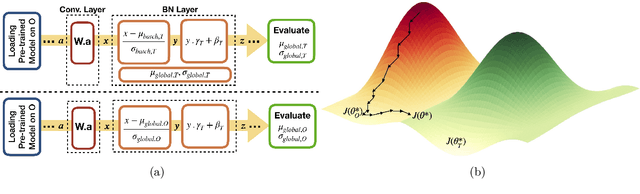
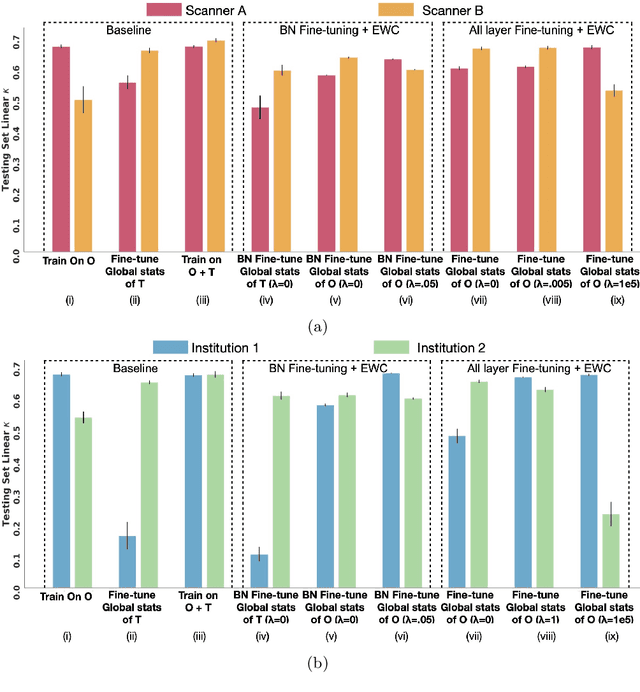
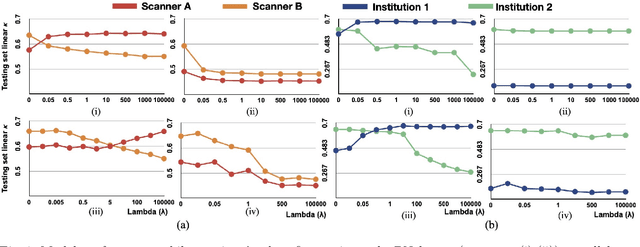
Abstract:Model brittleness is a key concern when deploying deep learning models in real-world medical settings. A model that has high performance at one institution may suffer a significant decline in performance when tested at other institutions. While pooling datasets from multiple institutions and retraining may provide a straightforward solution, it is often infeasible and may compromise patient privacy. An alternative approach is to fine-tune the model on subsequent institutions after training on the original institution. Notably, this approach degrades model performance at the original institution, a phenomenon known as catastrophic forgetting. In this paper, we develop an approach to address catastrophic forget-ting based on elastic weight consolidation combined with modulation of batch normalization statistics under two scenarios: first, for expanding the domain from one imaging system's data to another imaging system's, and second, for expanding the domain from a large multi-institutional dataset to another single institution dataset. We show that our approach outperforms several other state-of-the-art approaches and provide theoretical justification for the efficacy of batch normalization modulation. The results of this study are generally applicable to the deployment of any clinical deep learning model which requires domain expansion.
The unreasonable effectiveness of Batch-Norm statistics in addressing catastrophic forgetting across medical institutions
Nov 16, 2020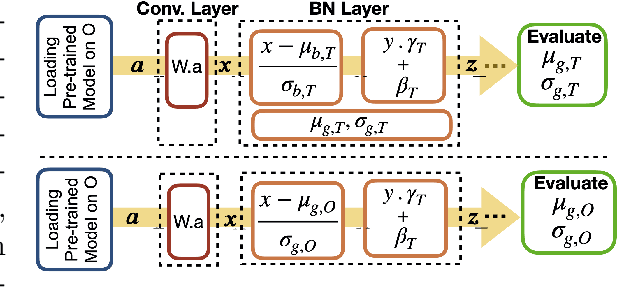
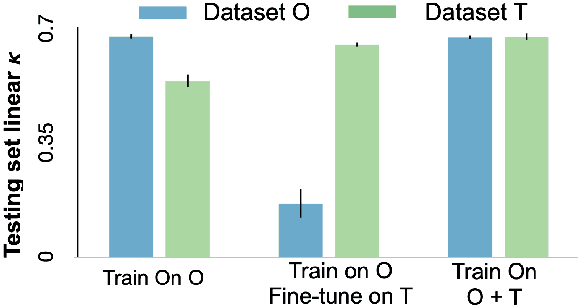
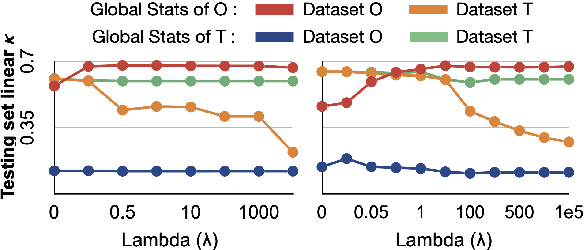
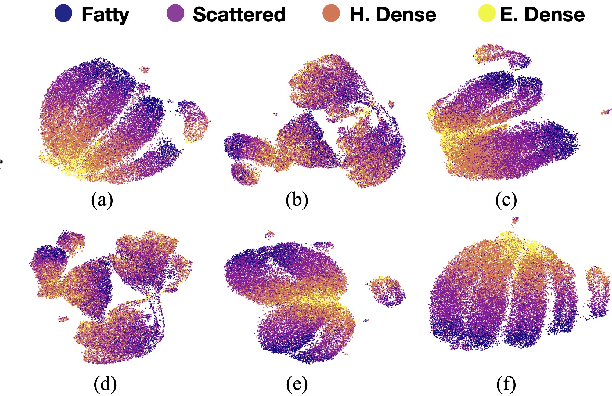
Abstract:Model brittleness is a primary concern when deploying deep learning models in medical settings owing to inter-institution variations, like patient demographics and intra-institution variation, such as multiple scanner types. While simply training on the combined datasets is fraught with data privacy limitations, fine-tuning the model on subsequent institutions after training it on the original institution results in a decrease in performance on the original dataset, a phenomenon called catastrophic forgetting. In this paper, we investigate trade-off between model refinement and retention of previously learned knowledge and subsequently address catastrophic forgetting for the assessment of mammographic breast density. More specifically, we propose a simple yet effective approach, adapting Elastic weight consolidation (EWC) using the global batch normalization (BN) statistics of the original dataset. The results of this study provide guidance for the deployment of clinical deep learning models where continuous learning is needed for domain expansion.
Towards Trainable Saliency Maps in Medical Imaging
Nov 15, 2020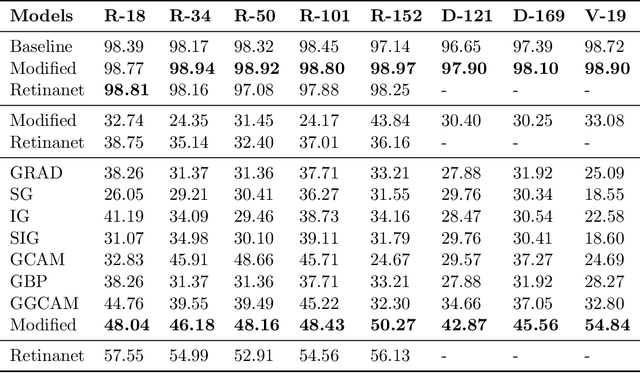
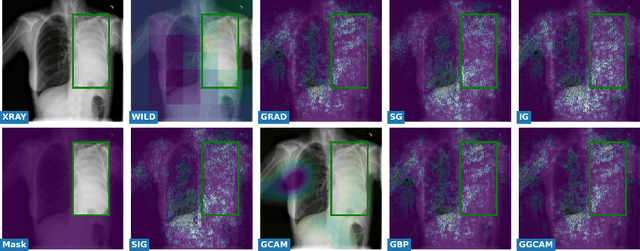
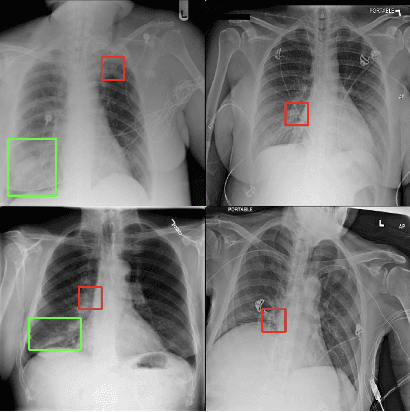
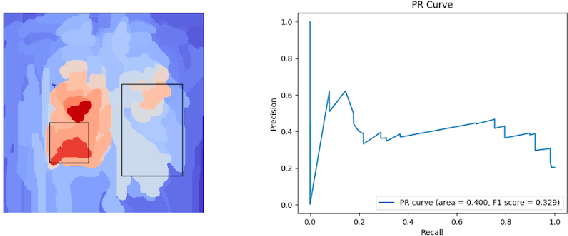
Abstract:While success of Deep Learning (DL) in automated diagnosis can be transformative to the medicinal practice especially for people with little or no access to doctors, its widespread acceptability is severely limited by inherent black-box decision making and unsafe failure modes. While saliency methods attempt to tackle this problem in non-medical contexts, their apriori explanations do not transfer well to medical usecases. With this study we validate a model design element agnostic to both architecture complexity and model task, and show how introducing this element gives an inherently self-explanatory model. We compare our results with state of the art non-trainable saliency maps on RSNA Pneumonia Dataset and demonstrate a much higher localization efficacy using our adopted technique. We also compare, with a fully supervised baseline and provide a reasonable alternative to it's high data labelling overhead. We further investigate the validity of our claims through qualitative evaluation from an expert reader.
An Autoencoder Based Approach to Simulate Sports Games
Jul 16, 2020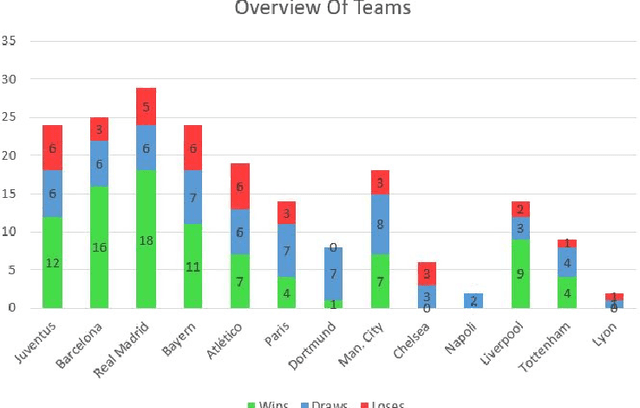

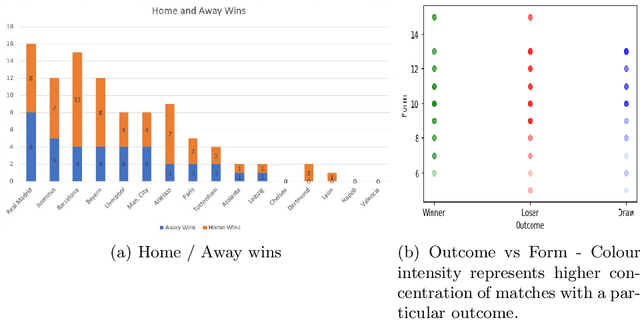
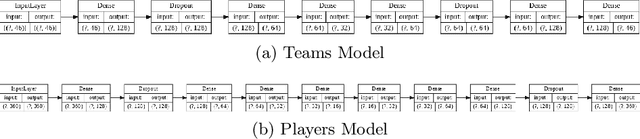
Abstract:Sports data has become widely available in the recent past. With the improvement of machine learning techniques, there have been attempts to use sports data to analyze not only the outcome of individual games but also to improve insights and strategies. The outbreak of COVID-19 has interrupted sports leagues globally, giving rise to increasing questions and speculations about the outcome of this season's leagues. What if the season was not interrupted and concluded normally? Which teams would end up winning trophies? Which players would perform the best? Which team would end their season on a high and which teams would fail to keep up with the pressure? We aim to tackle this problem and develop a solution. In this paper, we proposeUCLData, which is a dataset containing detailed information of UEFA Champions League games played over the past six years. We also propose a novel autoencoder based machine learning pipeline that can come up with a story on how the rest of the season will pan out.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge