Liangqiong Qu
Unveiling Implicit Advantage Symmetry: Why GRPO Struggles with Exploration and Difficulty Adaptation
Feb 05, 2026Abstract:Reinforcement Learning with Verifiable Rewards (RLVR), particularly GRPO, has become the standard for eliciting LLM reasoning. However, its efficiency in exploration and difficulty adaptation remains an open challenge. In this work, we argue that these bottlenecks stem from an implicit advantage symmetry inherent in Group Relative Advantage Estimation (GRAE). This symmetry induces two critical limitations: (i) at the group level, strict symmetry in weights between correct and incorrect trajectories leaves unsampled action logits unchanged, thereby hindering exploration of novel correct solution. (ii) at the sample level, the algorithm implicitly prioritizes medium-difficulty samples, remaining agnostic to the non-stationary demands of difficulty focus. Through controlled experiments, we reveal that this symmetric property is sub-optimal, yielding two pivotal insights: (i) asymmetrically suppressing the advantages of correct trajectories encourages essential exploration. (ii) learning efficiency is maximized by a curriculum-like transition-prioritizing simpler samples initially before gradually shifting to complex ones. Motivated by these findings, we propose Asymmetric GRAE (A-GRAE), which dynamically modulates exploration incentives and sample-difficulty focus. Experiments across seven benchmarks demonstrate that A-GRAE consistently improves GRPO and its variants across both LLMs and MLLMs.
FedVLMBench: Benchmarking Federated Fine-Tuning of Vision-Language Models
Jun 11, 2025Abstract:Vision-Language Models (VLMs) have demonstrated remarkable capabilities in cross-modal understanding and generation by integrating visual and textual information. While instruction tuning and parameter-efficient fine-tuning methods have substantially improved the generalization of VLMs, most existing approaches rely on centralized training, posing challenges for deployment in domains with strict privacy requirements like healthcare. Recent efforts have introduced Federated Learning (FL) into VLM fine-tuning to address these privacy concerns, yet comprehensive benchmarks for evaluating federated fine-tuning strategies, model architectures, and task generalization remain lacking. In this work, we present \textbf{FedVLMBench}, the first systematic benchmark for federated fine-tuning of VLMs. FedVLMBench integrates two mainstream VLM architectures (encoder-based and encoder-free), four fine-tuning strategies, five FL algorithms, six multimodal datasets spanning four cross-domain single-task scenarios and two cross-domain multitask settings, covering four distinct downstream task categories. Through extensive experiments, we uncover key insights into the interplay between VLM architectures, fine-tuning strategies, data heterogeneity, and multi-task federated optimization. Notably, we find that a 2-layer multilayer perceptron (MLP) connector with concurrent connector and LLM tuning emerges as the optimal configuration for encoder-based VLMs in FL. Furthermore, current FL methods exhibit significantly higher sensitivity to data heterogeneity in vision-centric tasks than text-centric ones, across both encoder-free and encoder-based VLM architectures. Our benchmark provides essential tools, datasets, and empirical guidance for the research community, offering a standardized platform to advance privacy-preserving, federated training of multimodal foundation models.
HSENet: Hybrid Spatial Encoding Network for 3D Medical Vision-Language Understanding
Jun 11, 2025Abstract:Automated 3D CT diagnosis empowers clinicians to make timely, evidence-based decisions by enhancing diagnostic accuracy and workflow efficiency. While multimodal large language models (MLLMs) exhibit promising performance in visual-language understanding, existing methods mainly focus on 2D medical images, which fundamentally limits their ability to capture complex 3D anatomical structures. This limitation often leads to misinterpretation of subtle pathologies and causes diagnostic hallucinations. In this paper, we present Hybrid Spatial Encoding Network (HSENet), a framework that exploits enriched 3D medical visual cues by effective visual perception and projection for accurate and robust vision-language understanding. Specifically, HSENet employs dual-3D vision encoders to perceive both global volumetric contexts and fine-grained anatomical details, which are pre-trained by dual-stage alignment with diagnostic reports. Furthermore, we propose Spatial Packer, an efficient multimodal projector that condenses high-resolution 3D spatial regions into a compact set of informative visual tokens via centroid-based compression. By assigning spatial packers with dual-3D vision encoders, HSENet can seamlessly perceive and transfer hybrid visual representations to LLM's semantic space, facilitating accurate diagnostic text generation. Experimental results demonstrate that our method achieves state-of-the-art performance in 3D language-visual retrieval (39.85% of R@100, +5.96% gain), 3D medical report generation (24.01% of BLEU-4, +8.01% gain), and 3D visual question answering (73.60% of Major Class Accuracy, +1.99% gain), confirming its effectiveness. Our code is available at https://github.com/YanzhaoShi/HSENet.
Distribution-aware Forgetting Compensation for Exemplar-Free Lifelong Person Re-identification
Apr 22, 2025Abstract:Lifelong Person Re-identification (LReID) suffers from a key challenge in preserving old knowledge while adapting to new information. The existing solutions include rehearsal-based and rehearsal-free methods to address this challenge. Rehearsal-based approaches rely on knowledge distillation, continuously accumulating forgetting during the distillation process. Rehearsal-free methods insufficiently learn the distribution of each domain, leading to forgetfulness over time. To solve these issues, we propose a novel Distribution-aware Forgetting Compensation (DAFC) model that explores cross-domain shared representation learning and domain-specific distribution integration without using old exemplars or knowledge distillation. We propose a Text-driven Prompt Aggregation (TPA) that utilizes text features to enrich prompt elements and guide the prompt model to learn fine-grained representations for each instance. This can enhance the differentiation of identity information and establish the foundation for domain distribution awareness. Then, Distribution-based Awareness and Integration (DAI) is designed to capture each domain-specific distribution by a dedicated expert network and adaptively consolidate them into a shared region in high-dimensional space. In this manner, DAI can consolidate and enhance cross-domain shared representation learning while alleviating catastrophic forgetting. Furthermore, we develop a Knowledge Consolidation Mechanism (KCM) that comprises instance-level discrimination and cross-domain consistency alignment strategies to facilitate model adaptive learning of new knowledge from the current domain and promote knowledge consolidation learning between acquired domain-specific distributions, respectively. Experimental results show that our DAFC outperforms state-of-the-art methods. Our code is available at https://github.com/LiuShiBen/DAFC.
TextSplat: Text-Guided Semantic Fusion for Generalizable Gaussian Splatting
Apr 13, 2025Abstract:Recent advancements in Generalizable Gaussian Splatting have enabled robust 3D reconstruction from sparse input views by utilizing feed-forward Gaussian Splatting models, achieving superior cross-scene generalization. However, while many methods focus on geometric consistency, they often neglect the potential of text-driven guidance to enhance semantic understanding, which is crucial for accurately reconstructing fine-grained details in complex scenes. To address this limitation, we propose TextSplat--the first text-driven Generalizable Gaussian Splatting framework. By employing a text-guided fusion of diverse semantic cues, our framework learns robust cross-modal feature representations that improve the alignment of geometric and semantic information, producing high-fidelity 3D reconstructions. Specifically, our framework employs three parallel modules to obtain complementary representations: the Diffusion Prior Depth Estimator for accurate depth information, the Semantic Aware Segmentation Network for detailed semantic information, and the Multi-View Interaction Network for refined cross-view features. Then, in the Text-Guided Semantic Fusion Module, these representations are integrated via the text-guided and attention-based feature aggregation mechanism, resulting in enhanced 3D Gaussian parameters enriched with detailed semantic cues. Experimental results on various benchmark datasets demonstrate improved performance compared to existing methods across multiple evaluation metrics, validating the effectiveness of our framework. The code will be publicly available.
Exploring the Vulnerabilities of Federated Learning: A Deep Dive into Gradient Inversion Attacks
Mar 13, 2025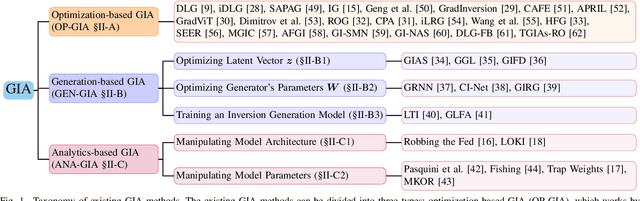
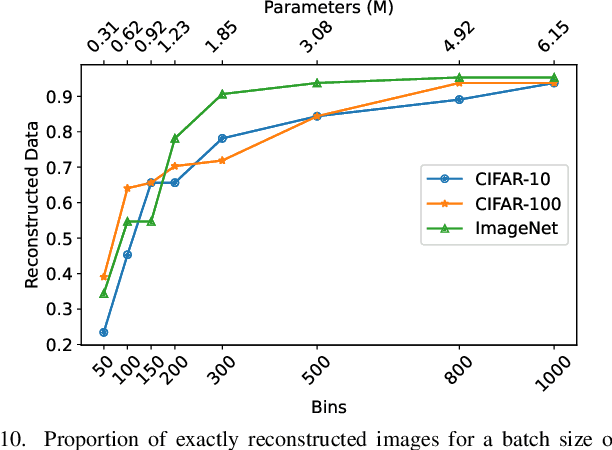
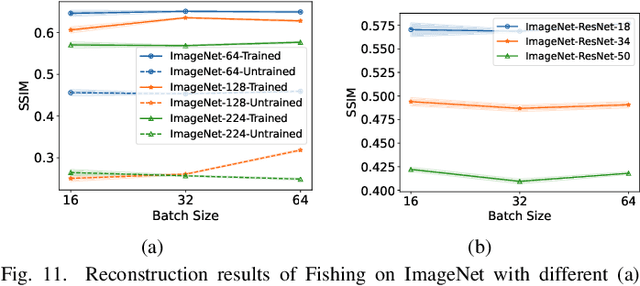
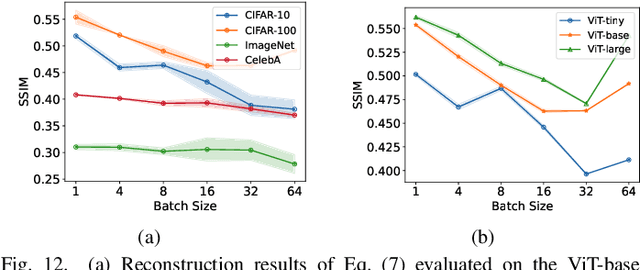
Abstract:Federated Learning (FL) has emerged as a promising privacy-preserving collaborative model training paradigm without sharing raw data. However, recent studies have revealed that private information can still be leaked through shared gradient information and attacked by Gradient Inversion Attacks (GIA). While many GIA methods have been proposed, a detailed analysis, evaluation, and summary of these methods are still lacking. Although various survey papers summarize existing privacy attacks in FL, few studies have conducted extensive experiments to unveil the effectiveness of GIA and their associated limiting factors in this context. To fill this gap, we first undertake a systematic review of GIA and categorize existing methods into three types, i.e., \textit{optimization-based} GIA (OP-GIA), \textit{generation-based} GIA (GEN-GIA), and \textit{analytics-based} GIA (ANA-GIA). Then, we comprehensively analyze and evaluate the three types of GIA in FL, providing insights into the factors that influence their performance, practicality, and potential threats. Our findings indicate that OP-GIA is the most practical attack setting despite its unsatisfactory performance, while GEN-GIA has many dependencies and ANA-GIA is easily detectable, making them both impractical. Finally, we offer a three-stage defense pipeline to users when designing FL frameworks and protocols for better privacy protection and share some future research directions from the perspectives of attackers and defenders that we believe should be pursued. We hope that our study can help researchers design more robust FL frameworks to defend against these attacks.
A New Federated Learning Framework Against Gradient Inversion Attacks
Dec 10, 2024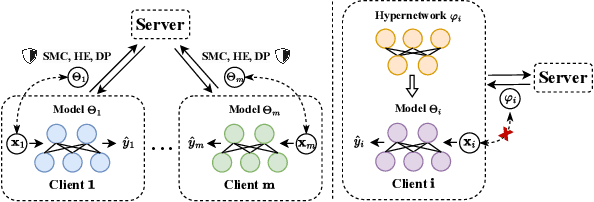
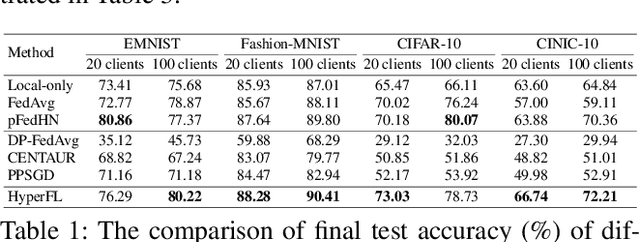
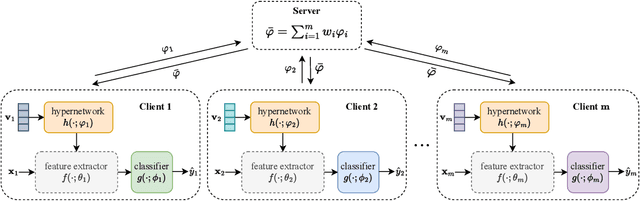
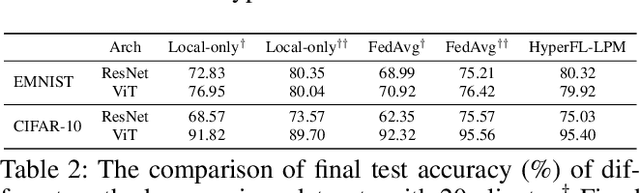
Abstract:Federated Learning (FL) aims to protect data privacy by enabling clients to collectively train machine learning models without sharing their raw data. However, recent studies demonstrate that information exchanged during FL is subject to Gradient Inversion Attacks (GIA) and, consequently, a variety of privacy-preserving methods have been integrated into FL to thwart such attacks, such as Secure Multi-party Computing (SMC), Homomorphic Encryption (HE), and Differential Privacy (DP). Despite their ability to protect data privacy, these approaches inherently involve substantial privacy-utility trade-offs. By revisiting the key to privacy exposure in FL under GIA, which lies in the frequent sharing of model gradients that contain private data, we take a new perspective by designing a novel privacy preserve FL framework that effectively ``breaks the direct connection'' between the shared parameters and the local private data to defend against GIA. Specifically, we propose a Hypernetwork Federated Learning (HyperFL) framework that utilizes hypernetworks to generate the parameters of the local model and only the hypernetwork parameters are uploaded to the server for aggregation. Theoretical analyses demonstrate the convergence rate of the proposed HyperFL, while extensive experimental results show the privacy-preserving capability and comparable performance of HyperFL. Code is available at https://github.com/Pengxin-Guo/HyperFL.
Selective Aggregation for Low-Rank Adaptation in Federated Learning
Oct 02, 2024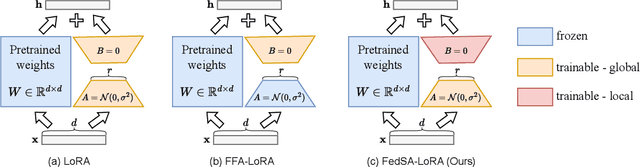
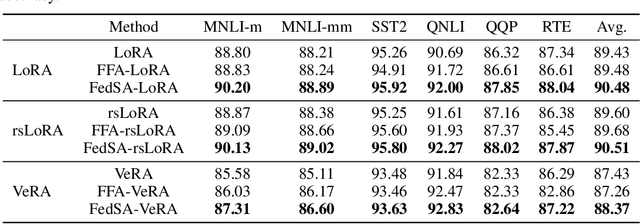
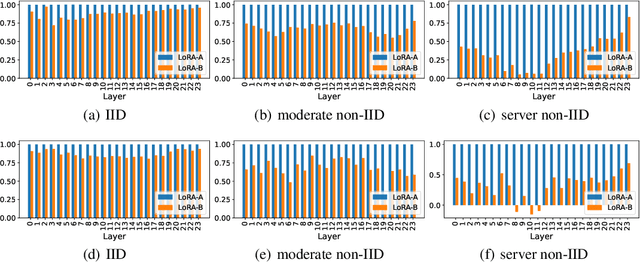
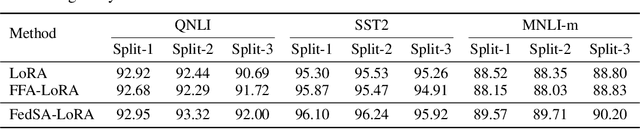
Abstract:We investigate LoRA in federated learning through the lens of the asymmetry analysis of the learned $A$ and $B$ matrices. In doing so, we uncover that $A$ matrices are responsible for learning general knowledge, while $B$ matrices focus on capturing client-specific knowledge. Based on this finding, we introduce Federated Share-A Low-Rank Adaptation (FedSA-LoRA), which employs two low-rank trainable matrices $A$ and $B$ to model the weight update, but only $A$ matrices are shared with the server for aggregation. Moreover, we delve into the relationship between the learned $A$ and $B$ matrices in other LoRA variants, such as rsLoRA and VeRA, revealing a consistent pattern. Consequently, we extend our FedSA-LoRA method to these LoRA variants, resulting in FedSA-rsLoRA and FedSA-VeRA. In this way, we establish a general paradigm for integrating LoRA with FL, offering guidance for future work on subsequent LoRA variants combined with FL. Extensive experimental results on natural language understanding and generation tasks demonstrate the effectiveness of the proposed method.
See Detail Say Clear: Towards Brain CT Report Generation via Pathological Clue-driven Representation Learning
Sep 29, 2024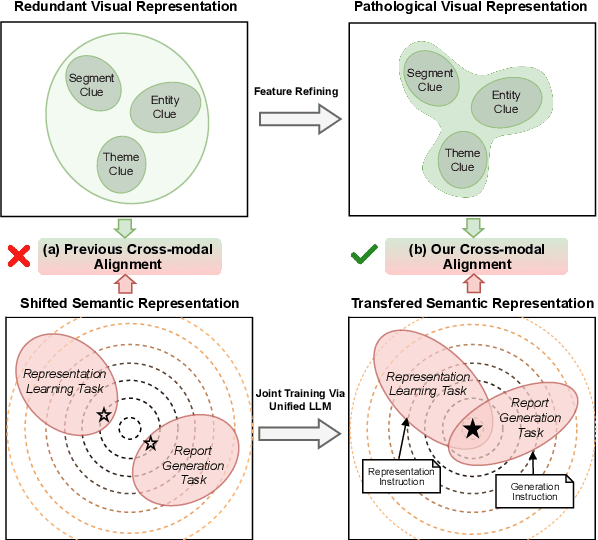
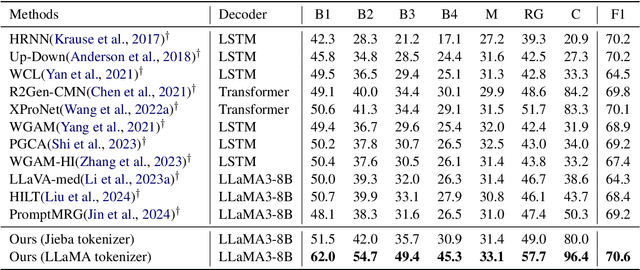
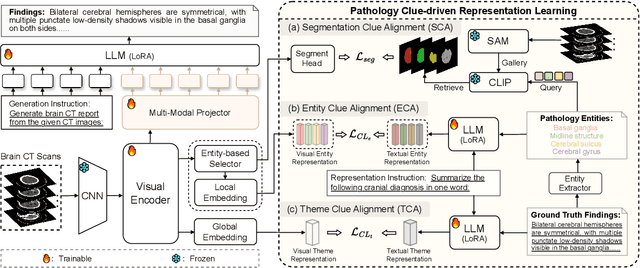
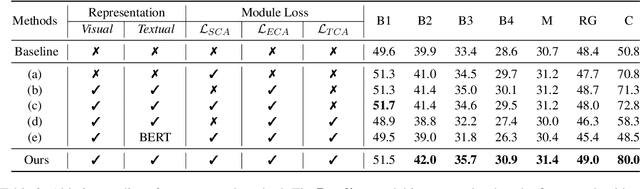
Abstract:Brain CT report generation is significant to aid physicians in diagnosing cranial diseases. Recent studies concentrate on handling the consistency between visual and textual pathological features to improve the coherence of report. However, there exist some challenges: 1) Redundant visual representing: Massive irrelevant areas in 3D scans distract models from representing salient visual contexts. 2) Shifted semantic representing: Limited medical corpus causes difficulties for models to transfer the learned textual representations to generative layers. This study introduces a Pathological Clue-driven Representation Learning (PCRL) model to build cross-modal representations based on pathological clues and naturally adapt them for accurate report generation. Specifically, we construct pathological clues from perspectives of segmented regions, pathological entities, and report themes, to fully grasp visual pathological patterns and learn cross-modal feature representations. To adapt the representations for the text generation task, we bridge the gap between representation learning and report generation by using a unified large language model (LLM) with task-tailored instructions. These crafted instructions enable the LLM to be flexibly fine-tuned across tasks and smoothly transfer the semantic representation for report generation. Experiments demonstrate that our method outperforms previous methods and achieves SoTA performance. Our code is available at https://github.com/Chauncey-Jheng/PCRL-MRG.
Unleashing the Potential of SAM for Medical Adaptation via Hierarchical Decoding
Mar 27, 2024Abstract:The Segment Anything Model (SAM) has garnered significant attention for its versatile segmentation abilities and intuitive prompt-based interface. However, its application in medical imaging presents challenges, requiring either substantial training costs and extensive medical datasets for full model fine-tuning or high-quality prompts for optimal performance. This paper introduces H-SAM: a prompt-free adaptation of SAM tailored for efficient fine-tuning of medical images via a two-stage hierarchical decoding procedure. In the initial stage, H-SAM employs SAM's original decoder to generate a prior probabilistic mask, guiding a more intricate decoding process in the second stage. Specifically, we propose two key designs: 1) A class-balanced, mask-guided self-attention mechanism addressing the unbalanced label distribution, enhancing image embedding; 2) A learnable mask cross-attention mechanism spatially modulating the interplay among different image regions based on the prior mask. Moreover, the inclusion of a hierarchical pixel decoder in H-SAM enhances its proficiency in capturing fine-grained and localized details. This approach enables SAM to effectively integrate learned medical priors, facilitating enhanced adaptation for medical image segmentation with limited samples. Our H-SAM demonstrates a 4.78% improvement in average Dice compared to existing prompt-free SAM variants for multi-organ segmentation using only 10% of 2D slices. Notably, without using any unlabeled data, H-SAM even outperforms state-of-the-art semi-supervised models relying on extensive unlabeled training data across various medical datasets. Our code is available at https://github.com/Cccccczh404/H-SAM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge