Qingyue Wei
Reasoning Within the Mind: Dynamic Multimodal Interleaving in Latent Space
Dec 17, 2025Abstract:Recent advancements in Multimodal Large Language Models (MLLMs) have significantly enhanced cross-modal understanding and reasoning by incorporating Chain-of-Thought (CoT) reasoning in the semantic space. Building upon this, recent studies extend the CoT mechanism to the visual modality, enabling models to integrate visual information during reasoning through external tools or explicit image generation. However, these methods remain dependent on explicit step-by-step reasoning, unstable perception-reasoning interaction and notable computational overhead. Inspired by human cognition, we posit that thinking unfolds not linearly but through the dynamic interleaving of reasoning and perception within the mind. Motivated by this perspective, we propose DMLR, a test-time Dynamic Multimodal Latent Reasoning framework that employs confidence-guided latent policy gradient optimization to refine latent think tokens for in-depth reasoning. Furthermore, a Dynamic Visual Injection Strategy is introduced, which retrieves the most relevant visual features at each latent think token and updates the set of best visual patches. The updated patches are then injected into latent think token to achieve dynamic visual-textual interleaving. Experiments across seven multimodal reasoning benchmarks and various model architectures demonstrate that DMLR significantly improves reasoning and perception performance while maintaining high inference efficiency.
RetinaRegNet: A Versatile Approach for Retinal Image Registration
Apr 24, 2024



Abstract:We introduce the RetinaRegNet model, which can achieve state-of-the-art performance across various retinal image registration tasks. RetinaRegNet does not require training on any retinal images. It begins by establishing point correspondences between two retinal images using image features derived from diffusion models. This process involves the selection of feature points from the moving image using the SIFT algorithm alongside random point sampling. For each selected feature point, a 2D correlation map is computed by assessing the similarity between the feature vector at that point and the feature vectors of all pixels in the fixed image. The pixel with the highest similarity score in the correlation map corresponds to the feature point in the moving image. To remove outliers in the estimated point correspondences, we first applied an inverse consistency constraint, followed by a transformation-based outlier detector. This method proved to outperform the widely used random sample consensus (RANSAC) outlier detector by a significant margin. To handle large deformations, we utilized a two-stage image registration framework. A homography transformation was used in the first stage and a more accurate third-order polynomial transformation was used in the second stage. The model's effectiveness was demonstrated across three retinal image datasets: color fundus images, fluorescein angiography images, and laser speckle flowgraphy images. RetinaRegNet outperformed current state-of-the-art methods in all three datasets. It was especially effective for registering image pairs with large displacement and scaling deformations. This innovation holds promise for various applications in retinal image analysis. Our code is publicly available at https://github.com/mirthAI/RetinaRegNet.
Unleashing the Potential of SAM for Medical Adaptation via Hierarchical Decoding
Mar 27, 2024Abstract:The Segment Anything Model (SAM) has garnered significant attention for its versatile segmentation abilities and intuitive prompt-based interface. However, its application in medical imaging presents challenges, requiring either substantial training costs and extensive medical datasets for full model fine-tuning or high-quality prompts for optimal performance. This paper introduces H-SAM: a prompt-free adaptation of SAM tailored for efficient fine-tuning of medical images via a two-stage hierarchical decoding procedure. In the initial stage, H-SAM employs SAM's original decoder to generate a prior probabilistic mask, guiding a more intricate decoding process in the second stage. Specifically, we propose two key designs: 1) A class-balanced, mask-guided self-attention mechanism addressing the unbalanced label distribution, enhancing image embedding; 2) A learnable mask cross-attention mechanism spatially modulating the interplay among different image regions based on the prior mask. Moreover, the inclusion of a hierarchical pixel decoder in H-SAM enhances its proficiency in capturing fine-grained and localized details. This approach enables SAM to effectively integrate learned medical priors, facilitating enhanced adaptation for medical image segmentation with limited samples. Our H-SAM demonstrates a 4.78% improvement in average Dice compared to existing prompt-free SAM variants for multi-organ segmentation using only 10% of 2D slices. Notably, without using any unlabeled data, H-SAM even outperforms state-of-the-art semi-supervised models relying on extensive unlabeled training data across various medical datasets. Our code is available at https://github.com/Cccccczh404/H-SAM.
3D TransUNet: Advancing Medical Image Segmentation through Vision Transformers
Oct 11, 2023Abstract:Medical image segmentation plays a crucial role in advancing healthcare systems for disease diagnosis and treatment planning. The u-shaped architecture, popularly known as U-Net, has proven highly successful for various medical image segmentation tasks. However, U-Net's convolution-based operations inherently limit its ability to model long-range dependencies effectively. To address these limitations, researchers have turned to Transformers, renowned for their global self-attention mechanisms, as alternative architectures. One popular network is our previous TransUNet, which leverages Transformers' self-attention to complement U-Net's localized information with the global context. In this paper, we extend the 2D TransUNet architecture to a 3D network by building upon the state-of-the-art nnU-Net architecture, and fully exploring Transformers' potential in both the encoder and decoder design. We introduce two key components: 1) A Transformer encoder that tokenizes image patches from a convolution neural network (CNN) feature map, enabling the extraction of global contexts, and 2) A Transformer decoder that adaptively refines candidate regions by utilizing cross-attention between candidate proposals and U-Net features. Our investigations reveal that different medical tasks benefit from distinct architectural designs. The Transformer encoder excels in multi-organ segmentation, where the relationship among organs is crucial. On the other hand, the Transformer decoder proves more beneficial for dealing with small and challenging segmented targets such as tumor segmentation. Extensive experiments showcase the significant potential of integrating a Transformer-based encoder and decoder into the u-shaped medical image segmentation architecture. TransUNet outperforms competitors in various medical applications.
Consistency-guided Meta-Learning for Bootstrapping Semi-Supervised Medical Image Segmentation
Jul 21, 2023



Abstract:Medical imaging has witnessed remarkable progress but usually requires a large amount of high-quality annotated data which is time-consuming and costly to obtain. To alleviate this burden, semi-supervised learning has garnered attention as a potential solution. In this paper, we present Meta-Learning for Bootstrapping Medical Image Segmentation (MLB-Seg), a novel method for tackling the challenge of semi-supervised medical image segmentation. Specifically, our approach first involves training a segmentation model on a small set of clean labeled images to generate initial labels for unlabeled data. To further optimize this bootstrapping process, we introduce a per-pixel weight mapping system that dynamically assigns weights to both the initialized labels and the model's own predictions. These weights are determined using a meta-process that prioritizes pixels with loss gradient directions closer to those of clean data, which is based on a small set of precisely annotated images. To facilitate the meta-learning process, we additionally introduce a consistency-based Pseudo Label Enhancement (PLE) scheme that improves the quality of the model's own predictions by ensembling predictions from various augmented versions of the same input. In order to improve the quality of the weight maps obtained through multiple augmentations of a single input, we introduce a mean teacher into the PLE scheme. This method helps to reduce noise in the weight maps and stabilize its generation process. Our extensive experimental results on public atrial and prostate segmentation datasets demonstrate that our proposed method achieves state-of-the-art results under semi-supervision. Our code is available at https://github.com/aijinrjinr/MLB-Seg.
Joint Graph Convolution for Analyzing Brain Structural and Functional Connectome
Oct 27, 2022Abstract:The white-matter (micro-)structural architecture of the brain promotes synchrony among neuronal populations, giving rise to richly patterned functional connections. A fundamental problem for systems neuroscience is determining the best way to relate structural and functional networks quantified by diffusion tensor imaging and resting-state functional MRI. As one of the state-of-the-art approaches for network analysis, graph convolutional networks (GCN) have been separately used to analyze functional and structural networks, but have not been applied to explore inter-network relationships. In this work, we propose to couple the two networks of an individual by adding inter-network edges between corresponding brain regions, so that the joint structure-function graph can be directly analyzed by a single GCN. The weights of inter-network edges are learnable, reflecting non-uniform structure-function coupling strength across the brain. We apply our Joint-GCN to predict age and sex of 662 participants from the public dataset of the National Consortium on Alcohol and Neurodevelopment in Adolescence (NCANDA) based on their functional and micro-structural white-matter networks. Our results support that the proposed Joint-GCN outperforms existing multi-modal graph learning approaches for analyzing structural and functional networks.
Label-Efficient Self-Supervised Federated Learning for Tackling Data Heterogeneity in Medical Imaging
May 17, 2022



Abstract:The curation of large-scale medical datasets from multiple institutions necessary for training deep learning models is challenged by the difficulty in sharing patient data with privacy-preserving. Federated learning (FL), a paradigm that enables privacy-protected collaborative learning among different institutions, is a promising solution to this challenge. However, FL generally suffers from performance deterioration due to heterogeneous data distributions across institutions and the lack of quality labeled data. In this paper, we present a robust and label-efficient self-supervised FL framework for medical image analysis. Specifically, we introduce a novel distributed self-supervised pre-training paradigm into the existing FL pipeline (i.e., pre-training the models directly on the decentralized target task datasets). Built upon the recent success of Vision Transformers, we employ masked image encoding tasks for self-supervised pre-training, to facilitate more effective knowledge transfer to downstream federated models. Extensive empirical results on simulated and real-world medical imaging federated datasets show that self-supervised pre-training largely benefits the robustness of federated models against various degrees of data heterogeneity. Notably, under severe data heterogeneity, our method, without relying on any additional pre-training data, achieves an improvement of 5.06%, 1.53% and 4.58% in test accuracy on retinal, dermatology and chest X-ray classification compared with the supervised baseline with ImageNet pre-training. Moreover, we show that our self-supervised FL algorithm generalizes well to out-of-distribution data and learns federated models more effectively in limited label scenarios, surpassing the supervised baseline by 10.36% and the semi-supervised FL method by 8.3% in test accuracy.
VerSe: A Vertebrae Labelling and Segmentation Benchmark
Jan 24, 2020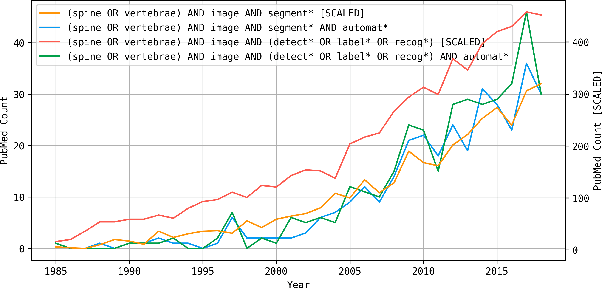
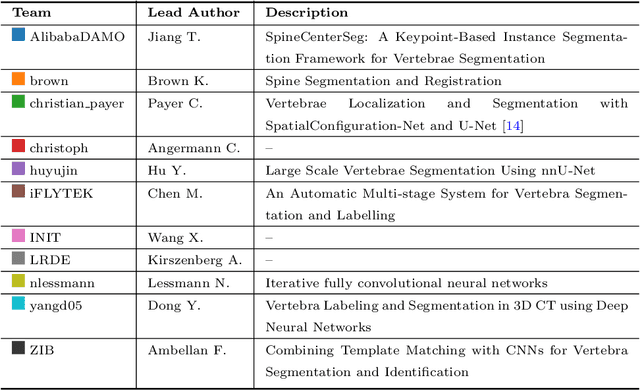
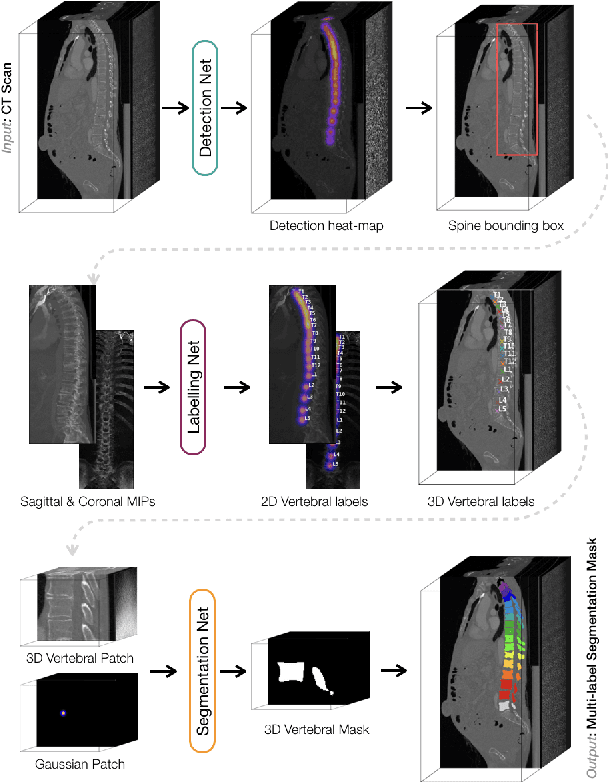
Abstract:In this paper we report the challenge set-up and results of the Large Scale Vertebrae Segmentation Challenge (VerSe) organized in conjunction with the MICCAI 2019. The challenge consisted of two tasks, vertebrae labelling and vertebrae segmentation. For this a total of 160 multidetector CT scan cohort closely resembling clinical setting was prepared and was annotated at a voxel-level by a human-machine hybrid algorithm. In this paper we also present the annotation protocol and the algorithm that aided the medical experts in the annotation process. Eleven fully automated algorithms were benchmarked on this data with the best performing algorithm achieving a vertebrae identification rate of 95% and a Dice coefficient of 90%. VerSe'19 is an open-call challenge at its image data along with the annotations and evaluation tools will continue to be publicly accessible through its online portal.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge