Alexander Valentinitsch
Institute of Signal Processing
Geometrically Inspired Kernel Machines for Collaborative Learning Beyond Gradient Descent
Jul 05, 2024Abstract:This paper develops a novel mathematical framework for collaborative learning by means of geometrically inspired kernel machines which includes statements on the bounds of generalisation and approximation errors, and sample complexity. For classification problems, this approach allows us to learn bounded geometric structures around given data points and hence solve the global model learning problem in an efficient way by exploiting convexity properties of the related optimisation problem in a Reproducing Kernel Hilbert Space (RKHS). In this way, we can reduce classification problems to determining the closest bounded geometric structure from a given data point. Further advantages that come with our solution is that our approach does not require clients to perform multiple epochs of local optimisation using stochastic gradient descent, nor require rounds of communication between client/server for optimising the global model. We highlight that numerous experiments have shown that the proposed method is a competitive alternative to the state-of-the-art.
VerSe: A Vertebrae Labelling and Segmentation Benchmark
Jan 24, 2020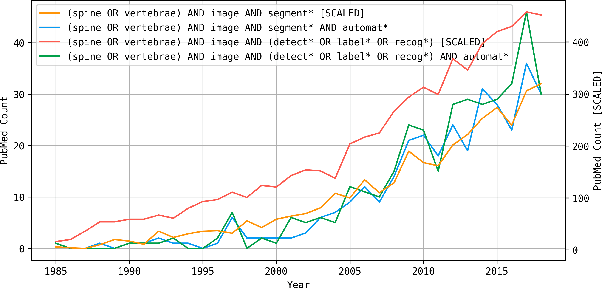
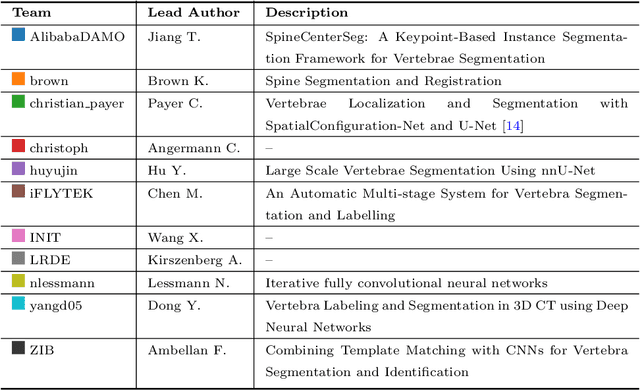
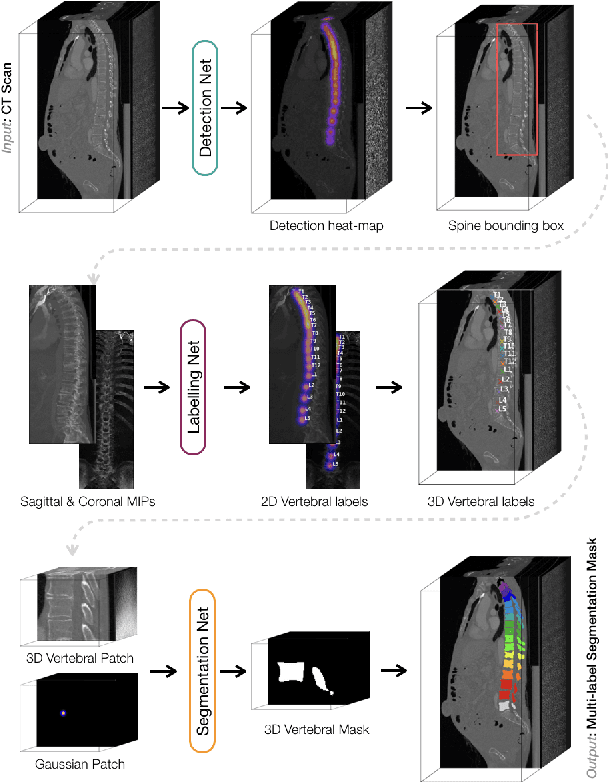
Abstract:In this paper we report the challenge set-up and results of the Large Scale Vertebrae Segmentation Challenge (VerSe) organized in conjunction with the MICCAI 2019. The challenge consisted of two tasks, vertebrae labelling and vertebrae segmentation. For this a total of 160 multidetector CT scan cohort closely resembling clinical setting was prepared and was annotated at a voxel-level by a human-machine hybrid algorithm. In this paper we also present the annotation protocol and the algorithm that aided the medical experts in the annotation process. Eleven fully automated algorithms were benchmarked on this data with the best performing algorithm achieving a vertebrae identification rate of 95% and a Dice coefficient of 90%. VerSe'19 is an open-call challenge at its image data along with the annotations and evaluation tools will continue to be publicly accessible through its online portal.
Probabilistic Point Cloud Reconstructions for Vertebral Shape Analysis
Aug 02, 2019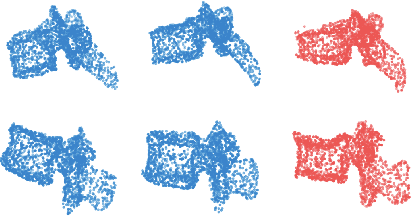
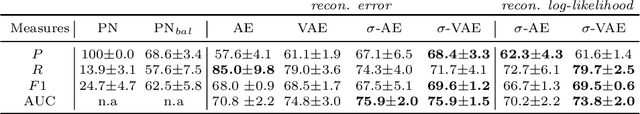
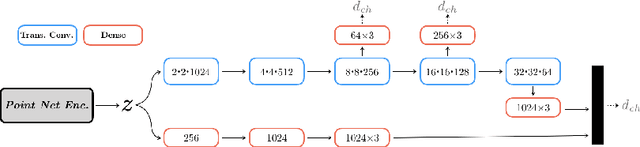
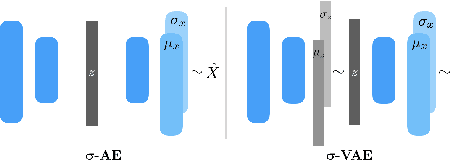
Abstract:We propose an auto-encoding network architecture for point clouds (PC) capable of extracting shape signatures without supervision. Building on this, we (i) design a loss function capable of modelling data variance on PCs which are unstructured, and (ii) regularise the latent space as in a variational auto-encoder, both of which increase the auto-encoders' descriptive capacity while making them probabilistic. Evaluating the reconstruction quality of our architectures, we employ them for detecting vertebral fractures without any supervision. By learning to efficiently reconstruct only healthy vertebrae, fractures are detected as anomalous reconstructions. Evaluating on a dataset containing $\sim$1500 vertebrae, we achieve area-under-ROC curve of $>$75%, without using intensity-based features.
Adversarially Learning a Local Anatomical Prior: Vertebrae Labelling with 2D reformations
Mar 03, 2019

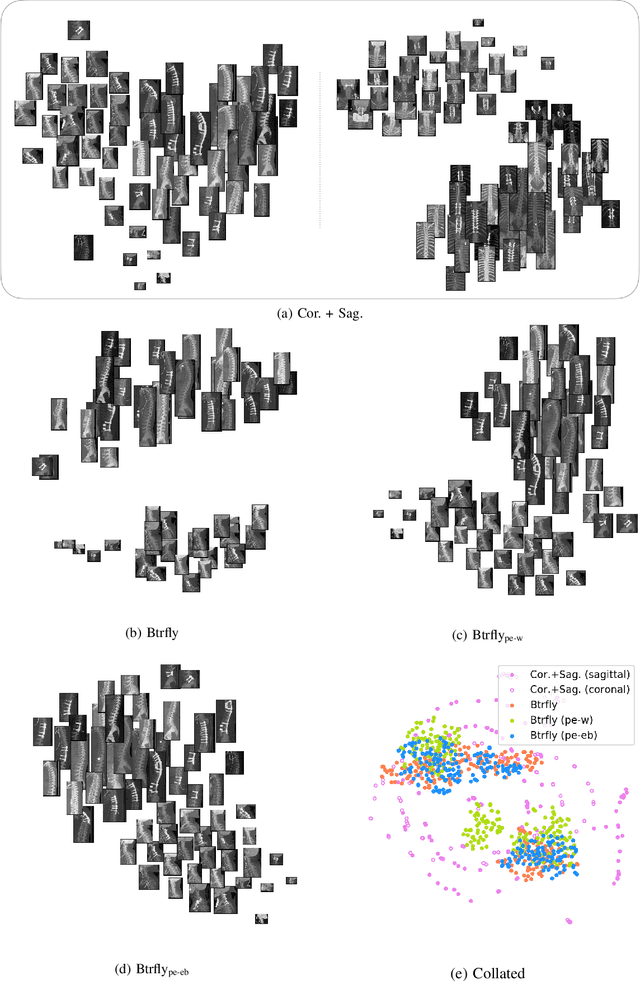
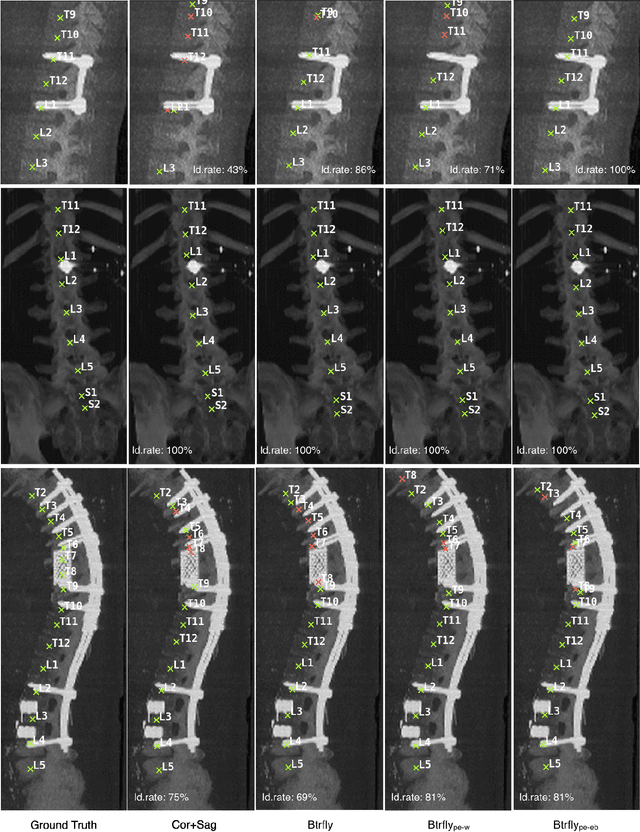
Abstract:Robust localisation and identification of vertebrae, jointly termed vertebrae labelling, in computed tomography (CT) images is an essential component of automated spine analysis. Current approaches for this task mostly work with 3D scans and are comprised of a sequence of multiple networks. Contrarily, our approach relies only on 2D reformations, enabling us to design an end-to-end trainable, standalone network. Our contribution includes: (1) Inspired by the workflow of human experts, a novel butterfly-shaped network architecture (termed Btrfly net) that efficiently combines information across sufficiently-informative sagittal and coronal reformations. (2) Two adversarial training regimes that encode an anatomical prior of the spine's shape into the Btrfly net, each enforcing the prior in a distinct manner. We evaluate our approach on a public benchmarking dataset of 302 CT scans achieving a performance comparable to state-of-art methods (identification rate of $>$88%) without any post-processing stages. Addressing its translation to clinical settings, an in-house dataset of 65 CT scans with a higher data variability is introduced, where we discuss refinements that render our approach robust to such scenarios.
Btrfly Net: Vertebrae Labelling with Energy-based Adversarial Learning of Local Spine Prior
Apr 04, 2018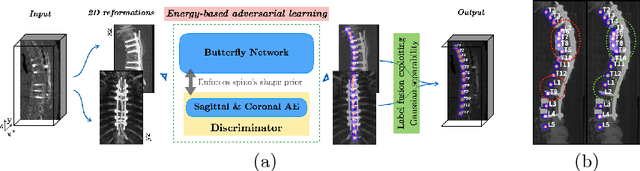
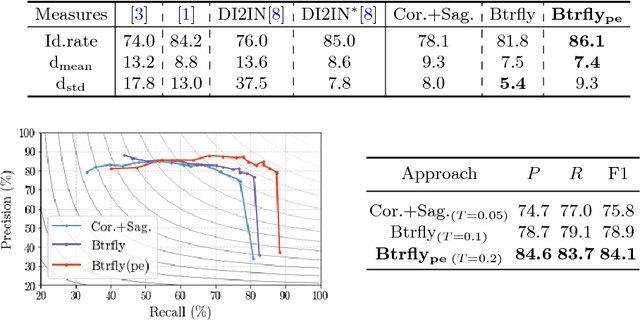
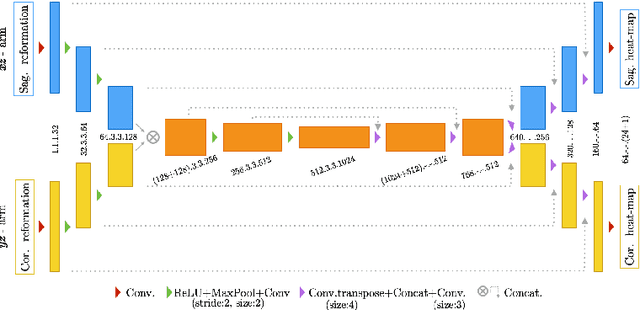
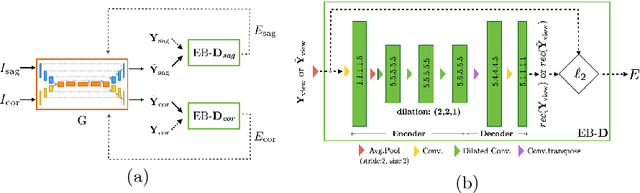
Abstract:Robust localisation and identification of vertebrae is an essential part of automated spine analysis. The contribution of this work to the task is two-fold: (1) Inspired by the human expert, we hypothesise that a sagittal and coronal reformation of the spine contain sufficient information for labelling the vertebrae. Thereby, we propose a butterfly-shaped network architecture (termed Btrfly Net) that efficiently combines the information across the reformations. (2) Underpinning the Btrfly net, we present an energy-based adversarial training regime that encodes the local spine structure as an anatomical prior into the network, thereby enabling it to achieve state-of-art performance in all standard metrics on a benchmark dataset of 302 scans without any post-processing during inference.
A Localisation-Segmentation Approach for Multi-label Annotation of Lumbar Vertebrae using Deep Nets
Mar 13, 2017



Abstract:Multi-class segmentation of vertebrae is a non-trivial task mainly due to the high correlation in the appearance of adjacent vertebrae. Hence, such a task calls for the consideration of both global and local context. Based on this motivation, we propose a two-staged approach that, given a computed tomography dataset of the spine, segments the five lumbar vertebrae and simultaneously labels them. The first stage employs a multi-layered perceptron performing non-linear regression for locating the lumbar region using the global context. The second stage, comprised of a fully-convolutional deep network, exploits the local context in the localised lumbar region to segment and label the lumbar vertebrae in one go. Aided with practical data augmentation for training, our approach is highly generalisable, capable of successfully segmenting both healthy and abnormal vertebrae (fractured and scoliotic spines). We consistently achieve an average Dice coefficient of over 90 percent on a publicly available dataset of the xVertSeg segmentation challenge of MICCAI 2016. This is particularly noteworthy because the xVertSeg dataset is beset with severe deformities in the form of vertebral fractures and scoliosis.
SurvivalNet: Predicting patient survival from diffusion weighted magnetic resonance images using cascaded fully convolutional and 3D convolutional neural networks
Feb 20, 2017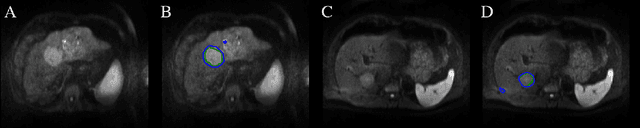

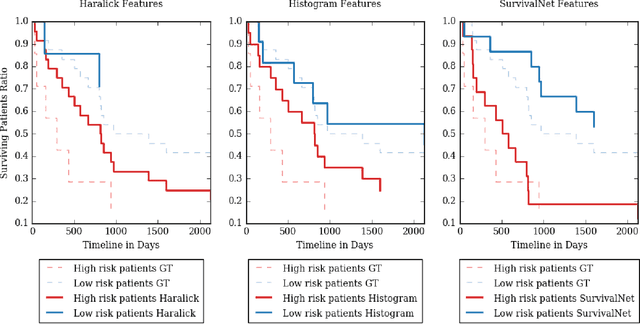
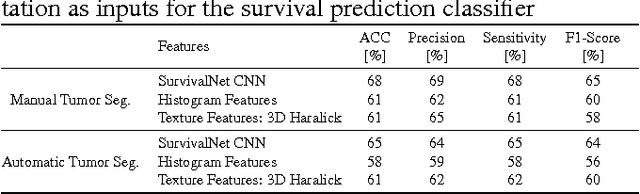
Abstract:Automatic non-invasive assessment of hepatocellular carcinoma (HCC) malignancy has the potential to substantially enhance tumor treatment strategies for HCC patients. In this work we present a novel framework to automatically characterize the malignancy of HCC lesions from DWI images. We predict HCC malignancy in two steps: As a first step we automatically segment HCC tumor lesions using cascaded fully convolutional neural networks (CFCN). A 3D neural network (SurvivalNet) then predicts the HCC lesions' malignancy from the HCC tumor segmentation. We formulate this task as a classification problem with classes being "low risk" and "high risk" represented by longer or shorter survival times than the median survival. We evaluated our method on DWI of 31 HCC patients. Our proposed framework achieves an end-to-end accuracy of 65% with a Dice score for the automatic lesion segmentation of 69% and an accuracy of 68% for tumor malignancy classification based on expert annotations. We compared the SurvivalNet to classical handcrafted features such as Histogram and Haralick and show experimentally that SurvivalNet outperforms the handcrafted features in HCC malignancy classification. End-to-end assessment of tumor malignancy based on our proposed fully automatic framework corresponds to assessment based on expert annotations with high significance (p>0.95).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge