Lequan Yu
Senior Member, IEEE
ExperienceWeaver: Optimizing Small-sample Experience Learning for LLM-based Clinical Text Improvement
Jan 31, 2026Abstract:Clinical text improvement is vital for healthcare efficiency but remains difficult due to limited high-quality data and the complex constraints of medical documentation. While Large Language Models (LLMs) show promise, current approaches struggle in small-sample settings: supervised fine-tuning is data-intensive and costly, while retrieval-augmented generation often provides superficial corrections without capturing the reasoning behind revisions. To address these limitations, we propose ExperienceWeaver, a hierarchical framework that shifts the focus from data retrieval to experience learning. Instead of simply recalling past examples, ExperienceWeaver distills noisy, multi-dimensional feedback into structured, actionable knowledge. Specifically, error-specific Tips and high-level Strategies. By injecting this distilled experience into an agentic pipeline, the model learns "how to revise" rather than just "what to revise". Extensive evaluations across four clinical datasets demonstrate that ExperienceWeaver consistently improves performance, surpassing state-of-the-art models such as Gemini-3 Pro in small-sample settings.
Augmenting Clinical Decision-Making with an Interactive and Interpretable AI Copilot: A Real-World User Study with Clinicians in Nephrology and Obstetrics
Jan 31, 2026Abstract:Clinician skepticism toward opaque AI hinders adoption in high-stakes healthcare. We present AICare, an interactive and interpretable AI copilot for collaborative clinical decision-making. By analyzing longitudinal electronic health records, AICare grounds dynamic risk predictions in scrutable visualizations and LLM-driven diagnostic recommendations. Through a within-subjects counterbalanced study with 16 clinicians across nephrology and obstetrics, we comprehensively evaluated AICare using objective measures (task completion time and error rate), subjective assessments (NASA-TLX, SUS, and confidence ratings), and semi-structured interviews. Our findings indicate AICare's reduced cognitive workload. Beyond performance metrics, qualitative analysis reveals that trust is actively constructed through verification, with interaction strategies diverging by expertise: junior clinicians used the system as cognitive scaffolding to structure their analysis, while experts engaged in adversarial verification to challenge the AI's logic. This work offers design implications for creating AI systems that function as transparent partners, accommodating diverse reasoning styles to augment rather than replace clinical judgment.
FDP: A Frequency-Decomposition Preprocessing Pipeline for Unsupervised Anomaly Detection in Brain MRI
Nov 17, 2025Abstract:Due to the diversity of brain anatomy and the scarcity of annotated data, supervised anomaly detection for brain MRI remains challenging, driving the development of unsupervised anomaly detection (UAD) approaches. Current UAD methods typically utilize artificially generated noise perturbations on healthy MRIs to train generative models for normal anatomy reconstruction, enabling anomaly detection via residual mapping. However, such simulated anomalies lack the biophysical fidelity and morphological complexity characteristic of true clinical lesions. To advance UAD in brain MRI, we conduct the first systematic frequency-domain analysis of pathological signatures, revealing two key properties: (1) anomalies exhibit unique frequency patterns distinguishable from normal anatomy, and (2) low-frequency signals maintain consistent representations across healthy scans. These insights motivate our Frequency-Decomposition Preprocessing (FDP) framework, the first UAD method to leverage frequency-domain reconstruction for simultaneous pathology suppression and anatomical preservation. FDP can integrate seamlessly with existing anomaly simulation techniques, consistently enhancing detection performance across diverse architectures while maintaining diagnostic fidelity. Experimental results demonstrate that FDP consistently improves anomaly detection performance when integrated with existing methods. Notably, FDP achieves a 17.63% increase in DICE score with LDM while maintaining robust improvements across multiple baselines. The code is available at https://github.com/ls1rius/MRI_FDP.
Variational Polya Tree
Oct 26, 2025Abstract:Density estimation is essential for generative modeling, particularly with the rise of modern neural networks. While existing methods capture complex data distributions, they often lack interpretability and uncertainty quantification. Bayesian nonparametric methods, especially the \polya tree, offer a robust framework that addresses these issues by accurately capturing function behavior over small intervals. Traditional techniques like Markov chain Monte Carlo (MCMC) face high computational complexity and scalability limitations, hindering the use of Bayesian nonparametric methods in deep learning. To tackle this, we introduce the variational \polya tree (VPT) model, which employs stochastic variational inference to compute posterior distributions. This model provides a flexible, nonparametric Bayesian prior that captures latent densities and works well with stochastic gradient optimization. We also leverage the joint distribution likelihood for a more precise variational posterior approximation than traditional mean-field methods. We evaluate the model performance on both real data and images, and demonstrate its competitiveness with other state-of-the-art deep density estimation methods. We also explore its ability in enhancing interpretability and uncertainty quantification. Code is available at https://github.com/howardchanth/var-polya-tree.
Amplifying Prominent Representations in Multimodal Learning via Variational Dirichlet Process
Oct 23, 2025Abstract:Developing effective multimodal fusion approaches has become increasingly essential in many real-world scenarios, such as health care and finance. The key challenge is how to preserve the feature expressiveness in each modality while learning cross-modal interactions. Previous approaches primarily focus on the cross-modal alignment, while over-emphasis on the alignment of marginal distributions of modalities may impose excess regularization and obstruct meaningful representations within each modality. The Dirichlet process (DP) mixture model is a powerful Bayesian non-parametric method that can amplify the most prominent features by its richer-gets-richer property, which allocates increasing weights to them. Inspired by this unique characteristic of DP, we propose a new DP-driven multimodal learning framework that automatically achieves an optimal balance between prominent intra-modal representation learning and cross-modal alignment. Specifically, we assume that each modality follows a mixture of multivariate Gaussian distributions and further adopt DP to calculate the mixture weights for all the components. This paradigm allows DP to dynamically allocate the contributions of features and select the most prominent ones, leveraging its richer-gets-richer property, thus facilitating multimodal feature fusion. Extensive experiments on several multimodal datasets demonstrate the superior performance of our model over other competitors. Ablation analysis further validates the effectiveness of DP in aligning modality distributions and its robustness to changes in key hyperparameters. Code is anonymously available at https://github.com/HKU-MedAI/DPMM.git
Large Material Gaussian Model for Relightable 3D Generation
Sep 26, 2025
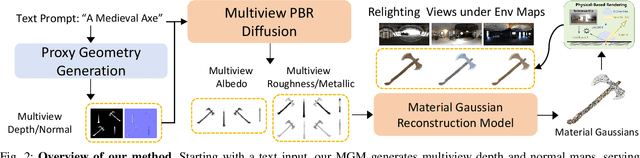
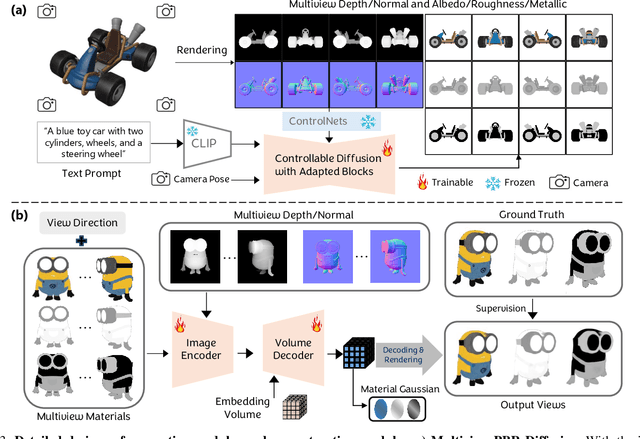

Abstract:The increasing demand for 3D assets across various industries necessitates efficient and automated methods for 3D content creation. Leveraging 3D Gaussian Splatting, recent large reconstruction models (LRMs) have demonstrated the ability to efficiently achieve high-quality 3D rendering by integrating multiview diffusion for generation and scalable transformers for reconstruction. However, existing models fail to produce the material properties of assets, which is crucial for realistic rendering in diverse lighting environments. In this paper, we introduce the Large Material Gaussian Model (MGM), a novel framework designed to generate high-quality 3D content with Physically Based Rendering (PBR) materials, ie, albedo, roughness, and metallic properties, rather than merely producing RGB textures with uncontrolled light baking. Specifically, we first fine-tune a new multiview material diffusion model conditioned on input depth and normal maps. Utilizing the generated multiview PBR images, we explore a Gaussian material representation that not only aligns with 2D Gaussian Splatting but also models each channel of the PBR materials. The reconstructed point clouds can then be rendered to acquire PBR attributes, enabling dynamic relighting by applying various ambient light maps. Extensive experiments demonstrate that the materials produced by our method not only exhibit greater visual appeal compared to baseline methods but also enhance material modeling, thereby enabling practical downstream rendering applications.
A Survey of Scientific Large Language Models: From Data Foundations to Agent Frontiers
Aug 28, 2025



Abstract:Scientific Large Language Models (Sci-LLMs) are transforming how knowledge is represented, integrated, and applied in scientific research, yet their progress is shaped by the complex nature of scientific data. This survey presents a comprehensive, data-centric synthesis that reframes the development of Sci-LLMs as a co-evolution between models and their underlying data substrate. We formulate a unified taxonomy of scientific data and a hierarchical model of scientific knowledge, emphasizing the multimodal, cross-scale, and domain-specific challenges that differentiate scientific corpora from general natural language processing datasets. We systematically review recent Sci-LLMs, from general-purpose foundations to specialized models across diverse scientific disciplines, alongside an extensive analysis of over 270 pre-/post-training datasets, showing why Sci-LLMs pose distinct demands -- heterogeneous, multi-scale, uncertainty-laden corpora that require representations preserving domain invariance and enabling cross-modal reasoning. On evaluation, we examine over 190 benchmark datasets and trace a shift from static exams toward process- and discovery-oriented assessments with advanced evaluation protocols. These data-centric analyses highlight persistent issues in scientific data development and discuss emerging solutions involving semi-automated annotation pipelines and expert validation. Finally, we outline a paradigm shift toward closed-loop systems where autonomous agents based on Sci-LLMs actively experiment, validate, and contribute to a living, evolving knowledge base. Collectively, this work provides a roadmap for building trustworthy, continually evolving artificial intelligence (AI) systems that function as a true partner in accelerating scientific discovery.
AssetDropper: Asset Extraction via Diffusion Models with Reward-Driven Optimization
Jun 06, 2025Abstract:Recent research on generative models has primarily focused on creating product-ready visual outputs; however, designers often favor access to standardized asset libraries, a domain that has yet to be significantly enhanced by generative capabilities. Although open-world scenes provide ample raw materials for designers, efficiently extracting high-quality, standardized assets remains a challenge. To address this, we introduce AssetDropper, the first framework designed to extract assets from reference images, providing artists with an open-world asset palette. Our model adeptly extracts a front view of selected subjects from input images, effectively handling complex scenarios such as perspective distortion and subject occlusion. We establish a synthetic dataset of more than 200,000 image-subject pairs and a real-world benchmark with thousands more for evaluation, facilitating the exploration of future research in downstream tasks. Furthermore, to ensure precise asset extraction that aligns well with the image prompts, we employ a pre-trained reward model to fulfill a closed-loop with feedback. We design the reward model to perform an inverse task that pastes the extracted assets back into the reference sources, which assists training with additional consistency and mitigates hallucination. Extensive experiments show that, with the aid of reward-driven optimization, AssetDropper achieves the state-of-the-art results in asset extraction. Project page: AssetDropper.github.io.
StyleAR: Customizing Multimodal Autoregressive Model for Style-Aligned Text-to-Image Generation
May 26, 2025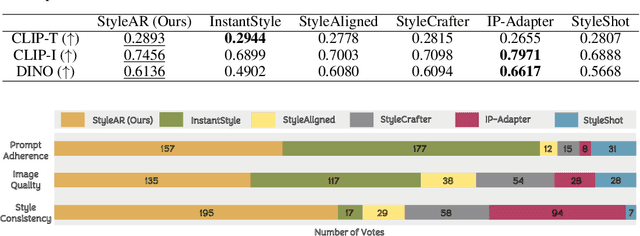
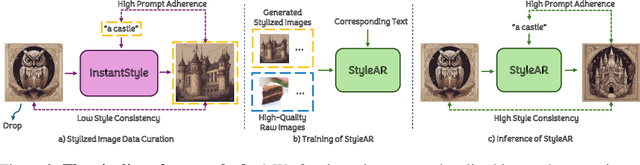
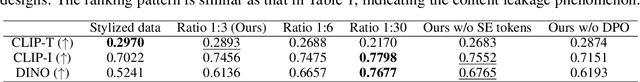
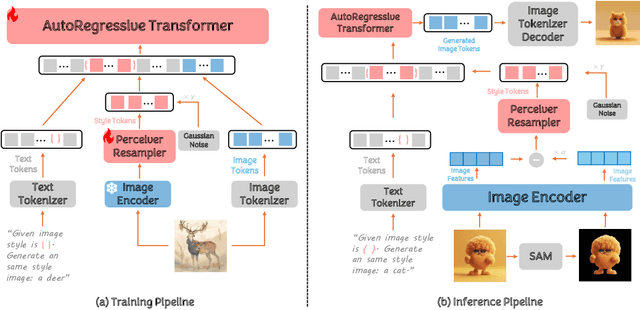
Abstract:In the current research landscape, multimodal autoregressive (AR) models have shown exceptional capabilities across various domains, including visual understanding and generation. However, complex tasks such as style-aligned text-to-image generation present significant challenges, particularly in data acquisition. In analogy to instruction-following tuning for image editing of AR models, style-aligned generation requires a reference style image and prompt, resulting in a text-image-to-image triplet where the output shares the style and semantics of the input. However, acquiring large volumes of such triplet data with specific styles is considerably more challenging than obtaining conventional text-to-image data used for training generative models. To address this issue, we propose StyleAR, an innovative approach that combines a specially designed data curation method with our proposed AR models to effectively utilize text-to-image binary data for style-aligned text-to-image generation. Our method synthesizes target stylized data using a reference style image and prompt, but only incorporates the target stylized image as the image modality to create high-quality binary data. To facilitate binary data training, we introduce a CLIP image encoder with a perceiver resampler that translates the image input into style tokens aligned with multimodal tokens in AR models and implement a style-enhanced token technique to prevent content leakage which is a common issue in previous work. Furthermore, we mix raw images drawn from large-scale text-image datasets with stylized images to enhance StyleAR's ability to extract richer stylistic features and ensure style consistency. Extensive qualitative and quantitative experiments demonstrate our superior performance.
Feature Preserving Shrinkage on Bayesian Neural Networks via the R2D2 Prior
May 23, 2025Abstract:Bayesian neural networks (BNNs) treat neural network weights as random variables, which aim to provide posterior uncertainty estimates and avoid overfitting by performing inference on the posterior weights. However, the selection of appropriate prior distributions remains a challenging task, and BNNs may suffer from catastrophic inflated variance or poor predictive performance when poor choices are made for the priors. Existing BNN designs apply different priors to weights, while the behaviours of these priors make it difficult to sufficiently shrink noisy signals or they are prone to overshrinking important signals in the weights. To alleviate this problem, we propose a novel R2D2-Net, which imposes the R^2-induced Dirichlet Decomposition (R2D2) prior to the BNN weights. The R2D2-Net can effectively shrink irrelevant coefficients towards zero, while preventing key features from over-shrinkage. To approximate the posterior distribution of weights more accurately, we further propose a variational Gibbs inference algorithm that combines the Gibbs updating procedure and gradient-based optimization. This strategy enhances stability and consistency in estimation when the variational objective involving the shrinkage parameters is non-convex. We also analyze the evidence lower bound (ELBO) and the posterior concentration rates from a theoretical perspective. Experiments on both natural and medical image classification and uncertainty estimation tasks demonstrate satisfactory performance of our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge