Tsai Hor Chan
Variational Polya Tree
Oct 26, 2025Abstract:Density estimation is essential for generative modeling, particularly with the rise of modern neural networks. While existing methods capture complex data distributions, they often lack interpretability and uncertainty quantification. Bayesian nonparametric methods, especially the \polya tree, offer a robust framework that addresses these issues by accurately capturing function behavior over small intervals. Traditional techniques like Markov chain Monte Carlo (MCMC) face high computational complexity and scalability limitations, hindering the use of Bayesian nonparametric methods in deep learning. To tackle this, we introduce the variational \polya tree (VPT) model, which employs stochastic variational inference to compute posterior distributions. This model provides a flexible, nonparametric Bayesian prior that captures latent densities and works well with stochastic gradient optimization. We also leverage the joint distribution likelihood for a more precise variational posterior approximation than traditional mean-field methods. We evaluate the model performance on both real data and images, and demonstrate its competitiveness with other state-of-the-art deep density estimation methods. We also explore its ability in enhancing interpretability and uncertainty quantification. Code is available at https://github.com/howardchanth/var-polya-tree.
Amplifying Prominent Representations in Multimodal Learning via Variational Dirichlet Process
Oct 23, 2025Abstract:Developing effective multimodal fusion approaches has become increasingly essential in many real-world scenarios, such as health care and finance. The key challenge is how to preserve the feature expressiveness in each modality while learning cross-modal interactions. Previous approaches primarily focus on the cross-modal alignment, while over-emphasis on the alignment of marginal distributions of modalities may impose excess regularization and obstruct meaningful representations within each modality. The Dirichlet process (DP) mixture model is a powerful Bayesian non-parametric method that can amplify the most prominent features by its richer-gets-richer property, which allocates increasing weights to them. Inspired by this unique characteristic of DP, we propose a new DP-driven multimodal learning framework that automatically achieves an optimal balance between prominent intra-modal representation learning and cross-modal alignment. Specifically, we assume that each modality follows a mixture of multivariate Gaussian distributions and further adopt DP to calculate the mixture weights for all the components. This paradigm allows DP to dynamically allocate the contributions of features and select the most prominent ones, leveraging its richer-gets-richer property, thus facilitating multimodal feature fusion. Extensive experiments on several multimodal datasets demonstrate the superior performance of our model over other competitors. Ablation analysis further validates the effectiveness of DP in aligning modality distributions and its robustness to changes in key hyperparameters. Code is anonymously available at https://github.com/HKU-MedAI/DPMM.git
Feature Preserving Shrinkage on Bayesian Neural Networks via the R2D2 Prior
May 23, 2025Abstract:Bayesian neural networks (BNNs) treat neural network weights as random variables, which aim to provide posterior uncertainty estimates and avoid overfitting by performing inference on the posterior weights. However, the selection of appropriate prior distributions remains a challenging task, and BNNs may suffer from catastrophic inflated variance or poor predictive performance when poor choices are made for the priors. Existing BNN designs apply different priors to weights, while the behaviours of these priors make it difficult to sufficiently shrink noisy signals or they are prone to overshrinking important signals in the weights. To alleviate this problem, we propose a novel R2D2-Net, which imposes the R^2-induced Dirichlet Decomposition (R2D2) prior to the BNN weights. The R2D2-Net can effectively shrink irrelevant coefficients towards zero, while preventing key features from over-shrinkage. To approximate the posterior distribution of weights more accurately, we further propose a variational Gibbs inference algorithm that combines the Gibbs updating procedure and gradient-based optimization. This strategy enhances stability and consistency in estimation when the variational objective involving the shrinkage parameters is non-convex. We also analyze the evidence lower bound (ELBO) and the posterior concentration rates from a theoretical perspective. Experiments on both natural and medical image classification and uncertainty estimation tasks demonstrate satisfactory performance of our method.
Democratizing Large Language Model-Based Graph Data Augmentation via Latent Knowledge Graphs
Feb 19, 2025Abstract:Data augmentation is necessary for graph representation learning due to the scarcity and noise present in graph data. Most of the existing augmentation methods overlook the context information inherited from the dataset as they rely solely on the graph structure for augmentation. Despite the success of some large language model-based (LLM) graph learning methods, they are mostly white-box which require access to the weights or latent features from the open-access LLMs, making them difficult to be democratized for everyone as existing LLMs are mostly closed-source for commercial considerations. To overcome these limitations, we propose a black-box context-driven graph data augmentation approach, with the guidance of LLMs -- DemoGraph. Leveraging the text prompt as context-related information, we task the LLM with generating knowledge graphs (KGs), which allow us to capture the structural interactions from the text outputs. We then design a dynamic merging schema to stochastically integrate the LLM-generated KGs into the original graph during training. To control the sparsity of the augmented graph, we further devise a granularity-aware prompting strategy and an instruction fine-tuning module, which seamlessly generates text prompts according to different granularity levels of the dataset. Extensive experiments on various graph learning tasks validate the effectiveness of our method over existing graph data augmentation methods. Notably, our approach excels in scenarios involving electronic health records (EHRs), which validates its maximal utilization of contextual knowledge, leading to enhanced predictive performance and interpretability.
Multi-task Heterogeneous Graph Learning on Electronic Health Records
Aug 14, 2024Abstract:Learning electronic health records (EHRs) has received emerging attention because of its capability to facilitate accurate medical diagnosis. Since the EHRs contain enriched information specifying complex interactions between entities, modeling EHRs with graphs is shown to be effective in practice. The EHRs, however, present a great degree of heterogeneity, sparsity, and complexity, which hamper the performance of most of the models applied to them. Moreover, existing approaches modeling EHRs often focus on learning the representations for a single task, overlooking the multi-task nature of EHR analysis problems and resulting in limited generalizability across different tasks. In view of these limitations, we propose a novel framework for EHR modeling, namely MulT-EHR (Multi-Task EHR), which leverages a heterogeneous graph to mine the complex relations and model the heterogeneity in the EHRs. To mitigate the large degree of noise, we introduce a denoising module based on the causal inference framework to adjust for severe confounding effects and reduce noise in the EHR data. Additionally, since our model adopts a single graph neural network for simultaneous multi-task prediction, we design a multi-task learning module to leverage the inter-task knowledge to regularize the training process. Extensive empirical studies on MIMIC-III and MIMIC-IV datasets validate that the proposed method consistently outperforms the state-of-the-art designs in four popular EHR analysis tasks -- drug recommendation, and predictions of the length of stay, mortality, and readmission. Thorough ablation studies demonstrate the robustness of our method upon variations to key components and hyperparameters.
cDP-MIL: Robust Multiple Instance Learning via Cascaded Dirichlet Process
Jul 16, 2024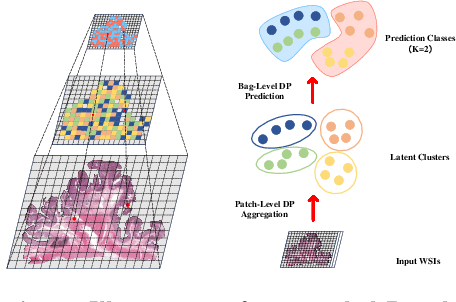
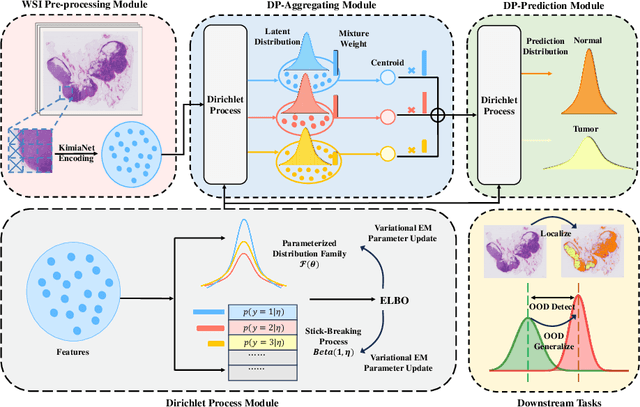
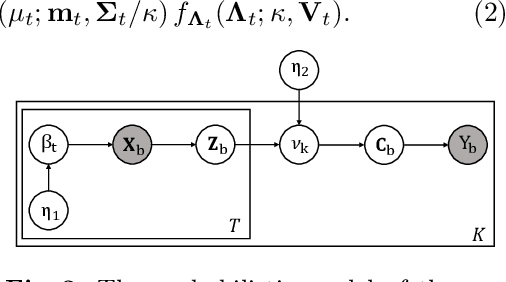
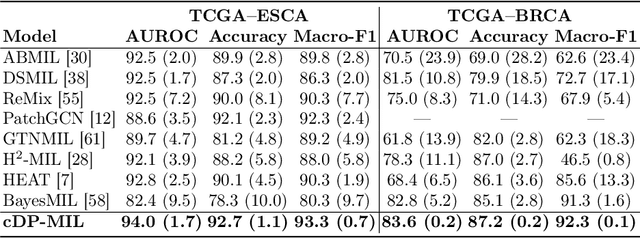
Abstract:Multiple instance learning (MIL) has been extensively applied to whole slide histopathology image (WSI) analysis. The existing aggregation strategy in MIL, which primarily relies on the first-order distance (e.g., mean difference) between instances, fails to accurately approximate the true feature distribution of each instance, leading to biased slide-level representations. Moreover, the scarcity of WSI observations easily leads to model overfitting, resulting in unstable testing performance and limited generalizability. To tackle these challenges, we propose a new Bayesian nonparametric framework for multiple instance learning, which adopts a cascade of Dirichlet processes (cDP) to incorporate the instance-to-bag characteristic of the WSIs. We perform feature aggregation based on the latent clusters formed by the Dirichlet process, which incorporates the covariances of the patch features and forms more representative clusters. We then perform bag-level prediction with another Dirichlet process model on the bags, which imposes a natural regularization on learning to prevent overfitting and enhance generalizability. Moreover, as a Bayesian nonparametric method, the cDP model can accurately generate posterior uncertainty, which allows for the detection of outlier samples and tumor localization. Extensive experiments on five WSI benchmarks validate the superior performance of our method, as well as its generalizability and ability to estimate uncertainties. Codes are available at https://github.com/HKU-MedAI/cDPMIL.
Adaptive Uncertainty Estimation via High-Dimensional Testing on Latent Representations
Oct 25, 2023Abstract:Uncertainty estimation aims to evaluate the confidence of a trained deep neural network. However, existing uncertainty estimation approaches rely on low-dimensional distributional assumptions and thus suffer from the high dimensionality of latent features. Existing approaches tend to focus on uncertainty on discrete classification probabilities, which leads to poor generalizability to uncertainty estimation for other tasks. Moreover, most of the literature requires seeing the out-of-distribution (OOD) data in the training for better estimation of uncertainty, which limits the uncertainty estimation performance in practice because the OOD data are typically unseen. To overcome these limitations, we propose a new framework using data-adaptive high-dimensional hypothesis testing for uncertainty estimation, which leverages the statistical properties of the feature representations. Our method directly operates on latent representations and thus does not require retraining the feature encoder under a modified objective. The test statistic relaxes the feature distribution assumptions to high dimensionality, and it is more discriminative to uncertainties in the latent representations. We demonstrate that encoding features with Bayesian neural networks can enhance testing performance and lead to more accurate uncertainty estimation. We further introduce a family-wise testing procedure to determine the optimal threshold of OOD detection, which minimizes the false discovery rate (FDR). Extensive experiments validate the satisfactory performance of our framework on uncertainty estimation and task-specific prediction over a variety of competitors. The experiments on the OOD detection task also show satisfactory performance of our method when the OOD data are unseen in the training. Codes are available at https://github.com/HKU-MedAI/bnn_uncertainty.
Source-Aware Embedding Training on Heterogeneous Information Networks
Jul 10, 2023Abstract:Heterogeneous information networks (HINs) have been extensively applied to real-world tasks, such as recommendation systems, social networks, and citation networks. While existing HIN representation learning methods can effectively learn the semantic and structural features in the network, little awareness was given to the distribution discrepancy of subgraphs within a single HIN. However, we find that ignoring such distribution discrepancy among subgraphs from multiple sources would hinder the effectiveness of graph embedding learning algorithms. This motivates us to propose SUMSHINE (Scalable Unsupervised Multi-Source Heterogeneous Information Network Embedding) -- a scalable unsupervised framework to align the embedding distributions among multiple sources of an HIN. Experimental results on real-world datasets in a variety of downstream tasks validate the performance of our method over the state-of-the-art heterogeneous information network embedding algorithms.
Histopathology Whole Slide Image Analysis with Heterogeneous Graph Representation Learning
Jul 09, 2023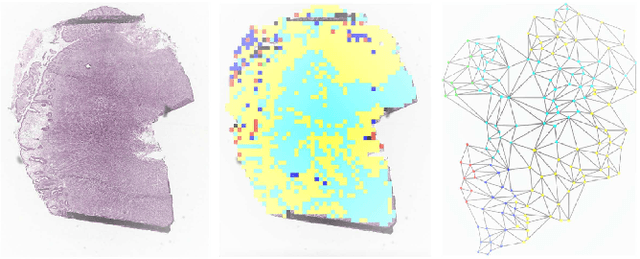
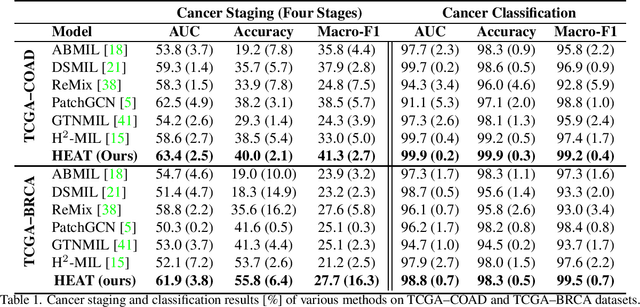
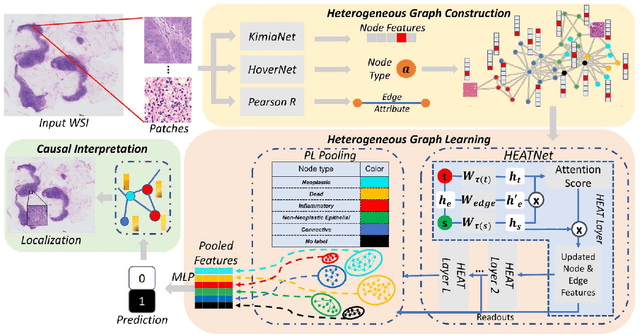
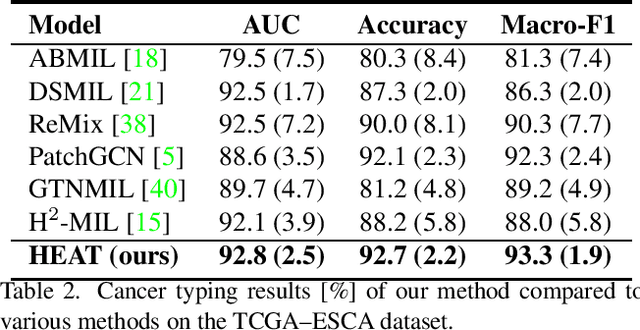
Abstract:Graph-based methods have been extensively applied to whole-slide histopathology image (WSI) analysis due to the advantage of modeling the spatial relationships among different entities. However, most of the existing methods focus on modeling WSIs with homogeneous graphs (e.g., with homogeneous node type). Despite their successes, these works are incapable of mining the complex structural relations between biological entities (e.g., the diverse interaction among different cell types) in the WSI. We propose a novel heterogeneous graph-based framework to leverage the inter-relationships among different types of nuclei for WSI analysis. Specifically, we formulate the WSI as a heterogeneous graph with "nucleus-type" attribute to each node and a semantic similarity attribute to each edge. We then present a new heterogeneous-graph edge attribute transformer (HEAT) to take advantage of the edge and node heterogeneity during massage aggregating. Further, we design a new pseudo-label-based semantic-consistent pooling mechanism to obtain graph-level features, which can mitigate the over-parameterization issue of conventional cluster-based pooling. Additionally, observing the limitations of existing association-based localization methods, we propose a causal-driven approach attributing the contribution of each node to improve the interpretability of our framework. Extensive experiments on three public TCGA benchmark datasets demonstrate that our framework outperforms the state-of-the-art methods with considerable margins on various tasks. Our codes are available at https://github.com/HKU-MedAI/WSI-HGNN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge