Xianhang Li
OpenVision 3: A Family of Unified Visual Encoder for Both Understanding and Generation
Jan 21, 2026Abstract:This paper presents a family of advanced vision encoder, named OpenVision 3, that learns a single, unified visual representation that can serve both image understanding and image generation. Our core architecture is simple: we feed VAE-compressed image latents to a ViT encoder and train its output to support two complementary roles. First, the encoder output is passed to the ViT-VAE decoder to reconstruct the original image, encouraging the representation to capture generative structure. Second, the same representation is optimized with contrastive learning and image-captioning objectives, strengthening semantic features. By jointly optimizing reconstruction- and semantics-driven signals in a shared latent space, the encoder learns representations that synergize and generalize well across both regimes. We validate this unified design through extensive downstream evaluations with the encoder frozen. For multimodal understanding, we plug the encoder into the LLaVA-1.5 framework: it performs comparably with a standard CLIP vision encoder (e.g., 62.4 vs 62.2 on SeedBench, and 83.7 vs 82.9 on POPE). For generation, we test it under the RAE framework: ours substantially surpasses the standard CLIP-based encoder (e.g., gFID: 1.89 vs 2.54 on ImageNet). We hope this work can spur future research on unified modeling.
OpenVision: A Fully-Open, Cost-Effective Family of Advanced Vision Encoders for Multimodal Learning
May 07, 2025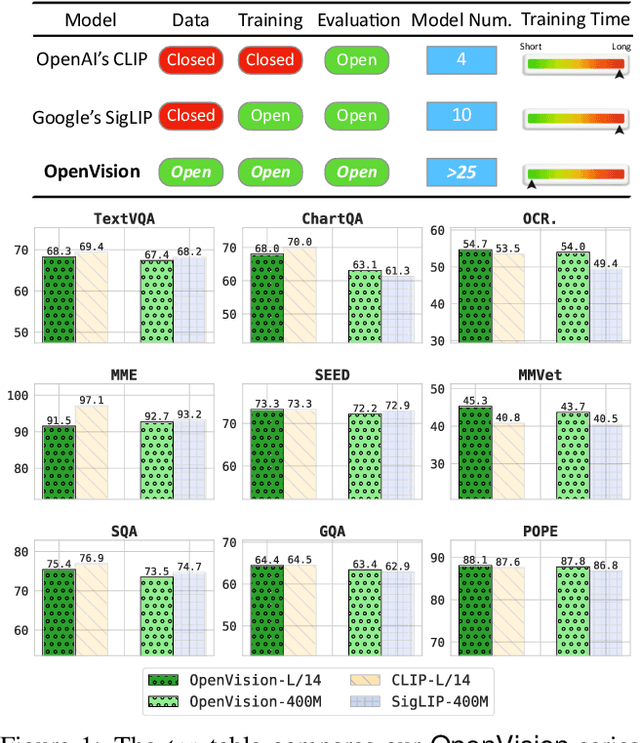
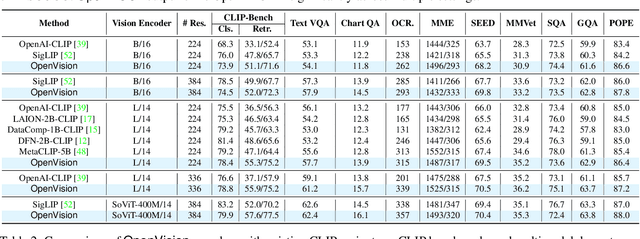
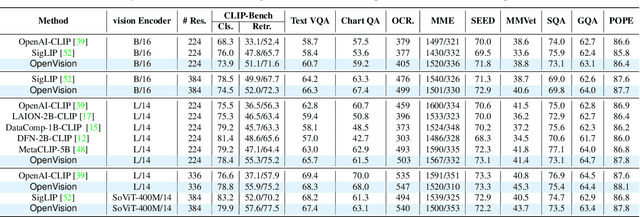
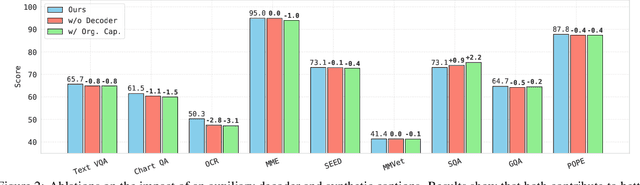
Abstract:OpenAI's CLIP, released in early 2021, have long been the go-to choice of vision encoder for building multimodal foundation models. Although recent alternatives such as SigLIP have begun to challenge this status quo, to our knowledge none are fully open: their training data remains proprietary and/or their training recipes are not released. This paper fills this gap with OpenVision, a fully-open, cost-effective family of vision encoders that match or surpass the performance of OpenAI's CLIP when integrated into multimodal frameworks like LLaVA. OpenVision builds on existing works -- e.g., CLIPS for training framework and Recap-DataComp-1B for training data -- while revealing multiple key insights in enhancing encoder quality and showcasing practical benefits in advancing multimodal models. By releasing vision encoders spanning from 5.9M to 632.1M parameters, OpenVision offers practitioners a flexible trade-off between capacity and efficiency in building multimodal models: larger models deliver enhanced multimodal performance, while smaller versions enable lightweight, edge-ready multimodal deployments.
CLIPS: An Enhanced CLIP Framework for Learning with Synthetic Captions
Nov 25, 2024



Abstract:Previous works show that noisy, web-crawled image-text pairs may limit vision-language pretraining like CLIP and propose learning with synthetic captions as a promising alternative. Our work continues this effort, introducing two simple yet effective designs to better leverage richly described synthetic captions. Firstly, by observing a strong inverse effect in learning with synthetic captions -- the short synthetic captions can generally lead to MUCH higher performance than full-length ones -- we therefore fed only partial synthetic captions to the text encoder. Secondly, we incorporate an autoregressive captioner to mimic the recaptioning process -- by conditioning on the paired image input and web-crawled text description, the captioner learns to predict the full-length synthetic caption generated by advanced MLLMs. Experiments show that our framework significantly improves zero-shot performance in cross-modal retrieval tasks, setting new SOTA results on MSCOCO and Flickr30K. Moreover, such trained vision encoders can enhance the visual capability of LLaVA, showing strong improvements on a range of MLLM benchmarks. Our project page is https://ucsc-vlaa.github.io/CLIPS/.
MedTrinity-25M: A Large-scale Multimodal Dataset with Multigranular Annotations for Medicine
Aug 06, 2024Abstract:This paper introduces MedTrinity-25M, a comprehensive, large-scale multimodal dataset for medicine, covering over 25 million images across 10 modalities, with multigranular annotations for more than 65 diseases. These enriched annotations encompass both global textual information, such as disease/lesion type, modality, region-specific descriptions, and inter-regional relationships, as well as detailed local annotations for regions of interest (ROIs), including bounding boxes, segmentation masks. Unlike existing approach which is limited by the availability of image-text pairs, we have developed the first automated pipeline that scales up multimodal data by generating multigranular visual and texual annotations (in the form of image-ROI-description triplets) without the need for any paired text descriptions. Specifically, data from over 90 different sources have been collected, preprocessed, and grounded using domain-specific expert models to identify ROIs related to abnormal regions. We then build a comprehensive knowledge base and prompt multimodal large language models to perform retrieval-augmented generation with the identified ROIs as guidance, resulting in multigranular texual descriptions. Compared to existing datasets, MedTrinity-25M provides the most enriched annotations, supporting a comprehensive range of multimodal tasks such as captioning and report generation, as well as vision-centric tasks like classification and segmentation. Pretraining on MedTrinity-25M, our model achieves state-of-the-art performance on VQA-RAD and PathVQA, surpassing both multimodal large language models and other representative SoTA approaches. This dataset can also be utilized to support large-scale pre-training of multimodal medical AI models, contributing to the development of future foundation models in the medical domain.
What If We Recaption Billions of Web Images with LLaMA-3?
Jun 12, 2024



Abstract:Web-crawled image-text pairs are inherently noisy. Prior studies demonstrate that semantically aligning and enriching textual descriptions of these pairs can significantly enhance model training across various vision-language tasks, particularly text-to-image generation. However, large-scale investigations in this area remain predominantly closed-source. Our paper aims to bridge this community effort, leveraging the powerful and \textit{open-sourced} LLaMA-3, a GPT-4 level LLM. Our recaptioning pipeline is simple: first, we fine-tune a LLaMA-3-8B powered LLaVA-1.5 and then employ it to recaption 1.3 billion images from the DataComp-1B dataset. Our empirical results confirm that this enhanced dataset, Recap-DataComp-1B, offers substantial benefits in training advanced vision-language models. For discriminative models like CLIP, we observe enhanced zero-shot performance in cross-modal retrieval tasks. For generative models like text-to-image Diffusion Transformers, the generated images exhibit a significant improvement in alignment with users' text instructions, especially in following complex queries. Our project page is https://www.haqtu.me/Recap-Datacomp-1B/
Autoregressive Pretraining with Mamba in Vision
Jun 11, 2024



Abstract:The vision community has started to build with the recently developed state space model, Mamba, as the new backbone for a range of tasks. This paper shows that Mamba's visual capability can be significantly enhanced through autoregressive pretraining, a direction not previously explored. Efficiency-wise, the autoregressive nature can well capitalize on the Mamba's unidirectional recurrent structure, enabling faster overall training speed compared to other training strategies like mask modeling. Performance-wise, autoregressive pretraining equips the Mamba architecture with markedly higher accuracy over its supervised-trained counterparts and, more importantly, successfully unlocks its scaling potential to large and even huge model sizes. For example, with autoregressive pretraining, a base-size Mamba attains 83.2\% ImageNet accuracy, outperforming its supervised counterpart by 2.0\%; our huge-size Mamba, the largest Vision Mamba to date, attains 85.0\% ImageNet accuracy (85.5\% when finetuned with $384\times384$ inputs), notably surpassing all other Mamba variants in vision. The code is available at \url{https://github.com/OliverRensu/ARM}.
Medical Vision Generalist: Unifying Medical Imaging Tasks in Context
Jun 08, 2024



Abstract:This study presents Medical Vision Generalist (MVG), the first foundation model capable of handling various medical imaging tasks -- such as cross-modal synthesis, image segmentation, denoising, and inpainting -- within a unified image-to-image generation framework. Specifically, MVG employs an in-context generation strategy that standardizes the handling of inputs and outputs as images. By treating these tasks as an image generation process conditioned on prompt image-label pairs and input images, this approach enables a flexible unification of various tasks, even those spanning different modalities and datasets. To capitalize on both local and global context, we design a hybrid method combining masked image modeling with autoregressive training for conditional image generation. This hybrid approach yields the most robust performance across all involved medical imaging tasks. To rigorously evaluate MVG's capabilities, we curated the first comprehensive generalist medical vision benchmark, comprising 13 datasets and spanning four imaging modalities (CT, MRI, X-ray, and micro-ultrasound). Our results consistently establish MVG's superior performance, outperforming existing vision generalists, such as Painter and LVM. Furthermore, MVG exhibits strong scalability, with its performance demonstrably improving when trained on a more diverse set of tasks, and can be effectively adapted to unseen datasets with only minimal task-specific samples. The code is available at \url{https://github.com/OliverRensu/MVG}.
Scaling White-Box Transformers for Vision
Jun 03, 2024



Abstract:CRATE, a white-box transformer architecture designed to learn compressed and sparse representations, offers an intriguing alternative to standard vision transformers (ViTs) due to its inherent mathematical interpretability. Despite extensive investigations into the scaling behaviors of language and vision transformers, the scalability of CRATE remains an open question which this paper aims to address. Specifically, we propose CRATE-$\alpha$, featuring strategic yet minimal modifications to the sparse coding block in the CRATE architecture design, and a light training recipe designed to improve the scalability of CRATE. Through extensive experiments, we demonstrate that CRATE-$\alpha$ can effectively scale with larger model sizes and datasets. For example, our CRATE-$\alpha$-B substantially outperforms the prior best CRATE-B model accuracy on ImageNet classification by 3.7%, achieving an accuracy of 83.2%. Meanwhile, when scaling further, our CRATE-$\alpha$-L obtains an ImageNet classification accuracy of 85.1%. More notably, these model performance improvements are achieved while preserving, and potentially even enhancing the interpretability of learned CRATE models, as we demonstrate through showing that the learned token representations of increasingly larger trained CRATE-$\alpha$ models yield increasingly higher-quality unsupervised object segmentation of images. The project page is https://rayjryang.github.io/CRATE-alpha/.
3D-TransUNet for Brain Metastases Segmentation in the BraTS2023 Challenge
Mar 23, 2024



Abstract:Segmenting brain tumors is complex due to their diverse appearances and scales. Brain metastases, the most common type of brain tumor, are a frequent complication of cancer. Therefore, an effective segmentation model for brain metastases must adeptly capture local intricacies to delineate small tumor regions while also integrating global context to understand broader scan features. The TransUNet model, which combines Transformer self-attention with U-Net's localized information, emerges as a promising solution for this task. In this report, we address brain metastases segmentation by training the 3D-TransUNet model on the Brain Tumor Segmentation (BraTS-METS) 2023 challenge dataset. Specifically, we explored two architectural configurations: the Encoder-only 3D-TransUNet, employing Transformers solely in the encoder, and the Decoder-only 3D-TransUNet, utilizing Transformers exclusively in the decoder. For Encoder-only 3D-TransUNet, we note that Masked-Autoencoder pre-training is required for a better initialization of the Transformer Encoder and thus accelerates the training process. We identify that the Decoder-only 3D-TransUNet model should offer enhanced efficacy in the segmentation of brain metastases, as indicated by our 5-fold cross-validation on the training set. However, our use of the Encoder-only 3D-TransUNet model already yield notable results, with an average lesion-wise Dice score of 59.8\% on the test set, securing second place in the BraTS-METS 2023 challenge.
Revisiting Adversarial Training at Scale
Jan 09, 2024Abstract:The machine learning community has witnessed a drastic change in the training pipeline, pivoted by those ''foundation models'' with unprecedented scales. However, the field of adversarial training is lagging behind, predominantly centered around small model sizes like ResNet-50, and tiny and low-resolution datasets like CIFAR-10. To bridge this transformation gap, this paper provides a modern re-examination with adversarial training, investigating its potential benefits when applied at scale. Additionally, we introduce an efficient and effective training strategy to enable adversarial training with giant models and web-scale data at an affordable computing cost. We denote this newly introduced framework as AdvXL. Empirical results demonstrate that AdvXL establishes new state-of-the-art robust accuracy records under AutoAttack on ImageNet-1K. For example, by training on DataComp-1B dataset, our AdvXL empowers a vanilla ViT-g model to substantially surpass the previous records of $l_{\infty}$-, $l_{2}$-, and $l_{1}$-robust accuracy by margins of 11.4%, 14.2% and 12.9%, respectively. This achievement posits AdvXL as a pioneering approach, charting a new trajectory for the efficient training of robust visual representations at significantly larger scales. Our code is available at https://github.com/UCSC-VLAA/AdvXL.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge