Yongyi Lu
Exploring Talking Head Models With Adjacent Frame Prior for Speech-Preserving Facial Expression Manipulation
Jan 19, 2026Abstract:Speech-Preserving Facial Expression Manipulation (SPFEM) is an innovative technique aimed at altering facial expressions in images and videos while retaining the original mouth movements. Despite advancements, SPFEM still struggles with accurate lip synchronization due to the complex interplay between facial expressions and mouth shapes. Capitalizing on the advanced capabilities of audio-driven talking head generation (AD-THG) models in synthesizing precise lip movements, our research introduces a novel integration of these models with SPFEM. We present a new framework, Talking Head Facial Expression Manipulation (THFEM), which utilizes AD-THG models to generate frames with accurately synchronized lip movements from audio inputs and SPFEM-altered images. However, increasing the number of frames generated by AD-THG models tends to compromise the realism and expression fidelity of the images. To counter this, we develop an adjacent frame learning strategy that finetunes AD-THG models to predict sequences of consecutive frames. This strategy enables the models to incorporate information from neighboring frames, significantly improving image quality during testing. Our extensive experimental evaluations demonstrate that this framework effectively preserves mouth shapes during expression manipulations, highlighting the substantial benefits of integrating AD-THG with SPFEM.
Learning Semantic-Aware Representation in Visual-Language Models for Multi-Label Recognition with Partial Labels
Dec 14, 2024



Abstract:Multi-label recognition with partial labels (MLR-PL), in which only some labels are known while others are unknown for each image, is a practical task in computer vision, since collecting large-scale and complete multi-label datasets is difficult in real application scenarios. Recently, vision language models (e.g. CLIP) have demonstrated impressive transferability to downstream tasks in data limited or label limited settings. However, current CLIP-based methods suffer from semantic confusion in MLR task due to the lack of fine-grained information in the single global visual and textual representation for all categories. In this work, we address this problem by introducing a semantic decoupling module and a category-specific prompt optimization method in CLIP-based framework. Specifically, the semantic decoupling module following the visual encoder learns category-specific feature maps by utilizing the semantic-guided spatial attention mechanism. Moreover, the category-specific prompt optimization method is introduced to learn text representations aligned with category semantics. Therefore, the prediction of each category is independent, which alleviate the semantic confusion problem. Extensive experiments on Microsoft COCO 2014 and Pascal VOC 2007 datasets demonstrate that the proposed framework significantly outperforms current state-of-art methods with a simpler model structure. Additionally, visual analysis shows that our method effectively separates information from different categories and achieves better performance compared to CLIP-based baseline method.
Exploiting Structural Consistency of Chest Anatomy for Unsupervised Anomaly Detection in Radiography Images
Mar 13, 2024



Abstract:Radiography imaging protocols focus on particular body regions, therefore producing images of great similarity and yielding recurrent anatomical structures across patients. Exploiting this structured information could potentially ease the detection of anomalies from radiography images. To this end, we propose a Simple Space-Aware Memory Matrix for In-painting and Detecting anomalies from radiography images (abbreviated as SimSID). We formulate anomaly detection as an image reconstruction task, consisting of a space-aware memory matrix and an in-painting block in the feature space. During the training, SimSID can taxonomize the ingrained anatomical structures into recurrent visual patterns, and in the inference, it can identify anomalies (unseen/modified visual patterns) from the test image. Our SimSID surpasses the state of the arts in unsupervised anomaly detection by +8.0%, +5.0%, and +9.9% AUC scores on ZhangLab, COVIDx, and CheXpert benchmark datasets, respectively. Code: https://github.com/MrGiovanni/SimSID
3D TransUNet: Advancing Medical Image Segmentation through Vision Transformers
Oct 11, 2023Abstract:Medical image segmentation plays a crucial role in advancing healthcare systems for disease diagnosis and treatment planning. The u-shaped architecture, popularly known as U-Net, has proven highly successful for various medical image segmentation tasks. However, U-Net's convolution-based operations inherently limit its ability to model long-range dependencies effectively. To address these limitations, researchers have turned to Transformers, renowned for their global self-attention mechanisms, as alternative architectures. One popular network is our previous TransUNet, which leverages Transformers' self-attention to complement U-Net's localized information with the global context. In this paper, we extend the 2D TransUNet architecture to a 3D network by building upon the state-of-the-art nnU-Net architecture, and fully exploring Transformers' potential in both the encoder and decoder design. We introduce two key components: 1) A Transformer encoder that tokenizes image patches from a convolution neural network (CNN) feature map, enabling the extraction of global contexts, and 2) A Transformer decoder that adaptively refines candidate regions by utilizing cross-attention between candidate proposals and U-Net features. Our investigations reveal that different medical tasks benefit from distinct architectural designs. The Transformer encoder excels in multi-organ segmentation, where the relationship among organs is crucial. On the other hand, the Transformer decoder proves more beneficial for dealing with small and challenging segmented targets such as tumor segmentation. Extensive experiments showcase the significant potential of integrating a Transformer-based encoder and decoder into the u-shaped medical image segmentation architecture. TransUNet outperforms competitors in various medical applications.
Boosting Dermatoscopic Lesion Segmentation via Diffusion Models with Visual and Textual Prompts
Oct 04, 2023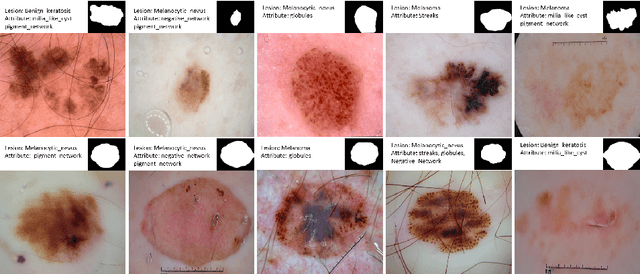
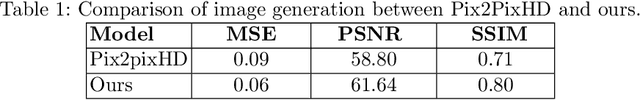
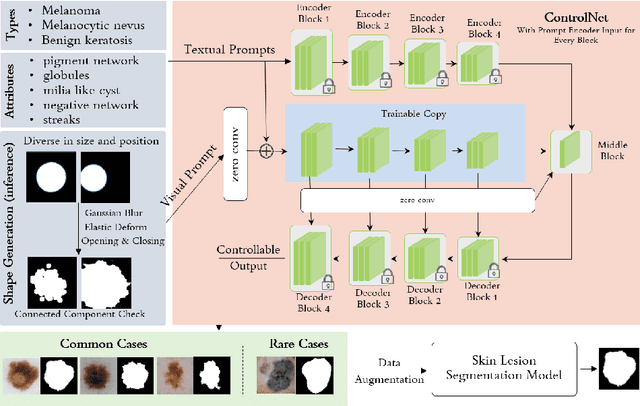
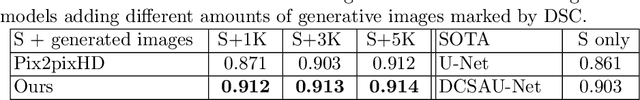
Abstract:Image synthesis approaches, e.g., generative adversarial networks, have been popular as a form of data augmentation in medical image analysis tasks. It is primarily beneficial to overcome the shortage of publicly accessible data and associated quality annotations. However, the current techniques often lack control over the detailed contents in generated images, e.g., the type of disease patterns, the location of lesions, and attributes of the diagnosis. In this work, we adapt the latest advance in the generative model, i.e., the diffusion model, with the added control flow using lesion-specific visual and textual prompts for generating dermatoscopic images. We further demonstrate the advantage of our diffusion model-based framework over the classical generation models in both the image quality and boosting the segmentation performance on skin lesions. It can achieve a 9% increase in the SSIM image quality measure and an over 5% increase in Dice coefficients over the prior arts.
Abdominal multi-organ segmentation in CT using Swinunter
Sep 28, 2023



Abstract:Abdominal multi-organ segmentation in computed tomography (CT) is crucial for many clinical applications including disease detection and treatment planning. Deep learning methods have shown unprecedented performance in this perspective. However, it is still quite challenging to accurately segment different organs utilizing a single network due to the vague boundaries of organs, the complex background, and the substantially different organ size scales. In this work we used make transformer-based model for training. It was found through previous years' competitions that basically all of the top 5 methods used CNN-based methods, which is likely due to the lack of data volume that prevents transformer-based methods from taking full advantage. The thousands of samples in this competition may enable the transformer-based model to have more excellent results. The results on the public validation set also show that the transformer-based model can achieve an acceptable result and inference time.
Spatial-Temporal Knowledge-Embedded Transformer for Video Scene Graph Generation
Sep 23, 2023



Abstract:Video scene graph generation (VidSGG) aims to identify objects in visual scenes and infer their relationships for a given video. It requires not only a comprehensive understanding of each object scattered on the whole scene but also a deep dive into their temporal motions and interactions. Inherently, object pairs and their relationships enjoy spatial co-occurrence correlations within each image and temporal consistency/transition correlations across different images, which can serve as prior knowledge to facilitate VidSGG model learning and inference. In this work, we propose a spatial-temporal knowledge-embedded transformer (STKET) that incorporates the prior spatial-temporal knowledge into the multi-head cross-attention mechanism to learn more representative relationship representations. Specifically, we first learn spatial co-occurrence and temporal transition correlations in a statistical manner. Then, we design spatial and temporal knowledge-embedded layers that introduce the multi-head cross-attention mechanism to fully explore the interaction between visual representation and the knowledge to generate spatial- and temporal-embedded representations, respectively. Finally, we aggregate these representations for each subject-object pair to predict the final semantic labels and their relationships. Extensive experiments show that STKET outperforms current competing algorithms by a large margin, e.g., improving the mR@50 by 8.1%, 4.7%, and 2.1% on different settings over current algorithms.
Learning to In-paint: Domain Adaptive Shape Completion for 3D Organ Segmentation
Aug 17, 2023Abstract:We aim at incorporating explicit shape information into current 3D organ segmentation models. Different from previous works, we formulate shape learning as an in-painting task, which is named Masked Label Mask Modeling (MLM). Through MLM, learnable mask tokens are fed into transformer blocks to complete the label mask of organ. To transfer MLM shape knowledge to target, we further propose a novel shape-aware self-distillation with both in-painting reconstruction loss and pseudo loss. Extensive experiments on five public organ segmentation datasets show consistent improvements over prior arts with at least 1.2 points gain in the Dice score, demonstrating the effectiveness of our method in challenging unsupervised domain adaptation scenarios including: (1) In-domain organ segmentation; (2) Unseen domain segmentation and (3) Unseen organ segmentation. We hope this work will advance shape analysis and geometric learning in medical imaging.
Data-Centric Diet: Effective Multi-center Dataset Pruning for Medical Image Segmentation
Aug 02, 2023Abstract:This paper seeks to address the dense labeling problems where a significant fraction of the dataset can be pruned without sacrificing much accuracy. We observe that, on standard medical image segmentation benchmarks, the loss gradient norm-based metrics of individual training examples applied in image classification fail to identify the important samples. To address this issue, we propose a data pruning method by taking into consideration the training dynamics on target regions using Dynamic Average Dice (DAD) score. To the best of our knowledge, we are among the first to address the data importance in dense labeling tasks in the field of medical image analysis, making the following contributions: (1) investigating the underlying causes with rigorous empirical analysis, and (2) determining effective data pruning approach in dense labeling problems. Our solution can be used as a strong yet simple baseline to select important examples for medical image segmentation with combined data sources.
Open-World Pose Transfer via Sequential Test-Time Adaption
Mar 20, 2023



Abstract:Pose transfer aims to transfer a given person into a specified posture, has recently attracted considerable attention. A typical pose transfer framework usually employs representative datasets to train a discriminative model, which is often violated by out-of-distribution (OOD) instances. Recently, test-time adaption (TTA) offers a feasible solution for OOD data by using a pre-trained model that learns essential features with self-supervision. However, those methods implicitly make an assumption that all test distributions have a unified signal that can be learned directly. In open-world conditions, the pose transfer task raises various independent signals: OOD appearance and skeleton, which need to be extracted and distributed in speciality. To address this point, we develop a SEquential Test-time Adaption (SETA). In the test-time phrase, SETA extracts and distributes external appearance texture by augmenting OOD data for self-supervised training. To make non-Euclidean similarity among different postures explicit, SETA uses the image representations derived from a person re-identification (Re-ID) model for similarity computation. By addressing implicit posture representation in the test-time sequentially, SETA greatly improves the generalization performance of current pose transfer models. In our experiment, we first show that pose transfer can be applied to open-world applications, including Tiktok reenactment and celebrity motion synthesis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge