Tiange Xiang
QuantiPhy: A Quantitative Benchmark Evaluating Physical Reasoning Abilities of Vision-Language Models
Dec 22, 2025Abstract:Understanding the physical world is essential for generalist AI agents. However, it remains unclear whether state-of-the-art vision perception models (e.g., large VLMs) can reason physical properties quantitatively. Existing evaluations are predominantly VQA-based and qualitative, offering limited insight into whether these models can infer the kinematic quantities of moving objects from video observations. To address this, we present QuantiPhy, the first benchmark designed to quantitatively measure a VLM's physical reasoning ability. Comprising more than 3.3K video-text instances with numerical ground truth, QuantiPhy evaluates a VLM's performance on estimating an object's size, velocity, and acceleration at a given timestamp, using one of these properties as an input prior. The benchmark standardizes prompts and scoring to assess numerical accuracy, enabling fair comparisons across models. Our experiments on state-of-the-art VLMs reveal a consistent gap between their qualitative plausibility and actual numerical correctness. We further provide an in-depth analysis of key factors like background noise, counterfactual priors, and strategic prompting and find that state-of-the-art VLMs lean heavily on pre-trained world knowledge rather than faithfully using the provided visual and textual inputs as references when reasoning kinematic properties quantitatively. QuantiPhy offers the first rigorous, scalable testbed to move VLMs beyond mere verbal plausibility toward a numerically grounded physical understanding.
ViBES: A Conversational Agent with Behaviorally-Intelligent 3D Virtual Body
Dec 16, 2025



Abstract:Human communication is inherently multimodal and social: words, prosody, and body language jointly carry intent. Yet most prior systems model human behavior as a translation task co-speech gesture or text-to-motion that maps a fixed utterance to motion clips-without requiring agentic decision-making about when to move, what to do, or how to adapt across multi-turn dialogue. This leads to brittle timing, weak social grounding, and fragmented stacks where speech, text, and motion are trained or inferred in isolation. We introduce ViBES (Voice in Behavioral Expression and Synchrony), a conversational 3D agent that jointly plans language and movement and executes dialogue-conditioned body actions. Concretely, ViBES is a speech-language-behavior (SLB) model with a mixture-of-modality-experts (MoME) backbone: modality-partitioned transformer experts for speech, facial expression, and body motion. The model processes interleaved multimodal token streams with hard routing by modality (parameters are split per expert), while sharing information through cross-expert attention. By leveraging strong pretrained speech-language models, the agent supports mixed-initiative interaction: users can speak, type, or issue body-action directives mid-conversation, and the system exposes controllable behavior hooks for streaming responses. We further benchmark on multi-turn conversation with automatic metrics of dialogue-motion alignment and behavior quality, and observe consistent gains over strong co-speech and text-to-motion baselines. ViBES goes beyond "speech-conditioned motion generation" toward agentic virtual bodies where language, prosody, and movement are jointly generated, enabling controllable, socially competent 3D interaction. Code and data will be made available at: ai.stanford.edu/~juze/ViBES/
Repurposing 2D Diffusion Models for 3D Shape Completion
Dec 16, 2025Abstract:We present a framework that adapts 2D diffusion models for 3D shape completion from incomplete point clouds. While text-to-image diffusion models have achieved remarkable success with abundant 2D data, 3D diffusion models lag due to the scarcity of high-quality 3D datasets and a persistent modality gap between 3D inputs and 2D latent spaces. To overcome these limitations, we introduce the Shape Atlas, a compact 2D representation of 3D geometry that (1) enables full utilization of the generative power of pretrained 2D diffusion models, and (2) aligns the modalities between the conditional input and output spaces, allowing more effective conditioning. This unified 2D formulation facilitates learning from limited 3D data and produces high-quality, detail-preserving shape completions. We validate the effectiveness of our results on the PCN and ShapeNet-55 datasets. Additionally, we show the downstream application of creating artist-created meshes from our completed point clouds, further demonstrating the practicality of our method.
SocialGen: Modeling Multi-Human Social Interaction with Language Models
Mar 28, 2025Abstract:Human interactions in everyday life are inherently social, involving engagements with diverse individuals across various contexts. Modeling these social interactions is fundamental to a wide range of real-world applications. In this paper, we introduce SocialGen, the first unified motion-language model capable of modeling interaction behaviors among varying numbers of individuals, to address this crucial yet challenging problem. Unlike prior methods that are limited to two-person interactions, we propose a novel social motion representation that supports tokenizing the motions of an arbitrary number of individuals and aligning them with the language space. This alignment enables the model to leverage rich, pretrained linguistic knowledge to better understand and reason about human social behaviors. To tackle the challenges of data scarcity, we curate a comprehensive multi-human interaction dataset, SocialX, enriched with textual annotations. Leveraging this dataset, we establish the first comprehensive benchmark for multi-human interaction tasks. Our method achieves state-of-the-art performance across motion-language tasks, setting a new standard for multi-human interaction modeling.
Repurposing 2D Diffusion Models with Gaussian Atlas for 3D Generation
Mar 20, 2025Abstract:Recent advances in text-to-image diffusion models have been driven by the increasing availability of paired 2D data. However, the development of 3D diffusion models has been hindered by the scarcity of high-quality 3D data, resulting in less competitive performance compared to their 2D counterparts. To address this challenge, we propose repurposing pre-trained 2D diffusion models for 3D object generation. We introduce Gaussian Atlas, a novel representation that utilizes dense 2D grids, enabling the fine-tuning of 2D diffusion models to generate 3D Gaussians. Our approach demonstrates successful transfer learning from a pre-trained 2D diffusion model to a 2D manifold flattened from 3D structures. To support model training, we compile GaussianVerse, a large-scale dataset comprising 205K high-quality 3D Gaussian fittings of various 3D objects. Our experimental results show that text-to-image diffusion models can be effectively adapted for 3D content generation, bridging the gap between 2D and 3D modeling.
Decoding Visual Experience and Mapping Semantics through Whole-Brain Analysis Using fMRI Foundation Models
Nov 11, 2024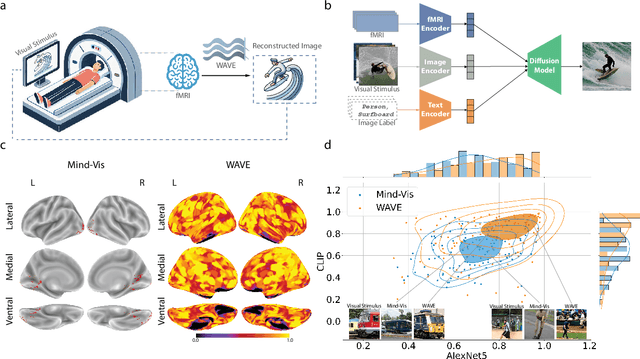
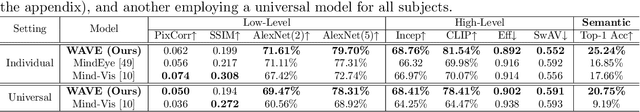


Abstract:Neural decoding, the process of understanding how brain activity corresponds to different stimuli, has been a primary objective in cognitive sciences. Over the past three decades, advancements in functional Magnetic Resonance Imaging and machine learning have greatly improved our ability to map visual stimuli to brain activity, especially in the visual cortex. Concurrently, research has expanded into decoding more complex processes like language and memory across the whole brain, utilizing techniques to handle greater variability and improve signal accuracy. We argue that "seeing" involves more than just mapping visual stimuli onto the visual cortex; it engages the entire brain, as various emotions and cognitive states can emerge from observing different scenes. In this paper, we develop algorithms to enhance our understanding of visual processes by incorporating whole-brain activation maps while individuals are exposed to visual stimuli. We utilize large-scale fMRI encoders and Image generative models pre-trained on large public datasets, which are then fine-tuned through Image-fMRI contrastive learning. Our models hence can decode visual experience across the entire cerebral cortex, surpassing the traditional confines of the visual cortex. We first compare our method with state-of-the-art approaches to decoding visual processing and show improved predictive semantic accuracy by 43%. A network ablation analysis suggests that beyond the visual cortex, the default mode network contributes most to decoding stimuli, in line with the proposed role of this network in sense-making and semantic processing. Additionally, we implemented zero-shot imagination decoding on an extra validation dataset, achieving a p-value of 0.0206 for mapping the reconstructed images and ground-truth text stimuli, which substantiates the model's capability to capture semantic meanings across various scenarios.
OccFusion: Rendering Occluded Humans with Generative Diffusion Priors
Jun 29, 2024Abstract:Most existing human rendering methods require every part of the human to be fully visible throughout the input video. However, this assumption does not hold in real-life settings where obstructions are common, resulting in only partial visibility of the human. Considering this, we present OccFusion, an approach that utilizes efficient 3D Gaussian splatting supervised by pretrained 2D diffusion models for efficient and high-fidelity human rendering. We propose a pipeline consisting of three stages. In the Initialization stage, complete human masks are generated from partial visibility masks. In the Optimization stage, 3D human Gaussians are optimized with additional supervision by Score-Distillation Sampling (SDS) to create a complete geometry of the human. Finally, in the Refinement stage, in-context inpainting is designed to further improve rendering quality on the less observed human body parts. We evaluate OccFusion on ZJU-MoCap and challenging OcMotion sequences and find that it achieves state-of-the-art performance in the rendering of occluded humans.
Exploiting Structural Consistency of Chest Anatomy for Unsupervised Anomaly Detection in Radiography Images
Mar 13, 2024



Abstract:Radiography imaging protocols focus on particular body regions, therefore producing images of great similarity and yielding recurrent anatomical structures across patients. Exploiting this structured information could potentially ease the detection of anomalies from radiography images. To this end, we propose a Simple Space-Aware Memory Matrix for In-painting and Detecting anomalies from radiography images (abbreviated as SimSID). We formulate anomaly detection as an image reconstruction task, consisting of a space-aware memory matrix and an in-painting block in the feature space. During the training, SimSID can taxonomize the ingrained anatomical structures into recurrent visual patterns, and in the inference, it can identify anomalies (unseen/modified visual patterns) from the test image. Our SimSID surpasses the state of the arts in unsupervised anomaly detection by +8.0%, +5.0%, and +9.9% AUC scores on ZhangLab, COVIDx, and CheXpert benchmark datasets, respectively. Code: https://github.com/MrGiovanni/SimSID
Wild2Avatar: Rendering Humans Behind Occlusions
Dec 31, 2023Abstract:Rendering the visual appearance of moving humans from occluded monocular videos is a challenging task. Most existing research renders 3D humans under ideal conditions, requiring a clear and unobstructed scene. Those methods cannot be used to render humans in real-world scenes where obstacles may block the camera's view and lead to partial occlusions. In this work, we present Wild2Avatar, a neural rendering approach catered for occluded in-the-wild monocular videos. We propose occlusion-aware scene parameterization for decoupling the scene into three parts - occlusion, human, and background. Additionally, extensive objective functions are designed to help enforce the decoupling of the human from both the occlusion and the background and to ensure the completeness of the human model. We verify the effectiveness of our approach with experiments on in-the-wild videos.
Rendering Humans from Object-Occluded Monocular Videos
Aug 08, 2023



Abstract:3D understanding and rendering of moving humans from monocular videos is a challenging task. Despite recent progress, the task remains difficult in real-world scenarios, where obstacles may block the camera view and cause partial occlusions in the captured videos. Existing methods cannot handle such defects due to two reasons. First, the standard rendering strategy relies on point-point mapping, which could lead to dramatic disparities between the visible and occluded areas of the body. Second, the naive direct regression approach does not consider any feasibility criteria (ie, prior information) for rendering under occlusions. To tackle the above drawbacks, we present OccNeRF, a neural rendering method that achieves better rendering of humans in severely occluded scenes. As direct solutions to the two drawbacks, we propose surface-based rendering by integrating geometry and visibility priors. We validate our method on both simulated and real-world occlusions and demonstrate our method's superiority.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge