Mahmut Yurt
ALN-P3: Unified Language Alignment for Perception, Prediction, and Planning in Autonomous Driving
May 21, 2025Abstract:Recent advances have explored integrating large language models (LLMs) into end-to-end autonomous driving systems to enhance generalization and interpretability. However, most existing approaches are limited to either driving performance or vision-language reasoning, making it difficult to achieve both simultaneously. In this paper, we propose ALN-P3, a unified co-distillation framework that introduces cross-modal alignment between "fast" vision-based autonomous driving systems and "slow" language-driven reasoning modules. ALN-P3 incorporates three novel alignment mechanisms: Perception Alignment (P1A), Prediction Alignment (P2A), and Planning Alignment (P3A), which explicitly align visual tokens with corresponding linguistic outputs across the full perception, prediction, and planning stack. All alignment modules are applied only during training and incur no additional costs during inference. Extensive experiments on four challenging benchmarks-nuScenes, Nu-X, TOD3Cap, and nuScenes QA-demonstrate that ALN-P3 significantly improves both driving decisions and language reasoning, achieving state-of-the-art results.
LTDA-Drive: LLMs-guided Generative Models based Long-tail Data Augmentation for Autonomous Driving
May 21, 2025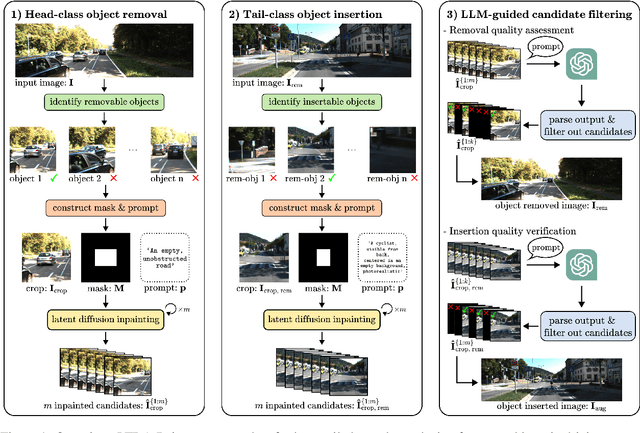
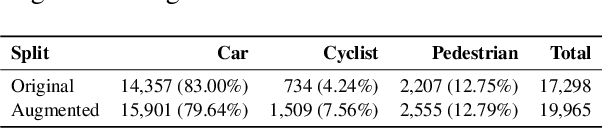
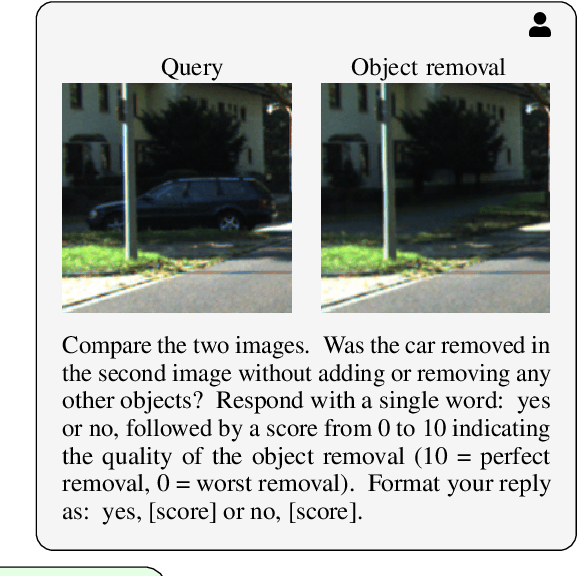
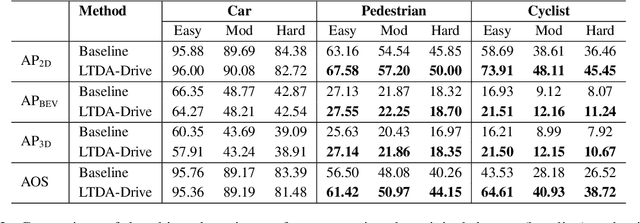
Abstract:3D perception plays an essential role for improving the safety and performance of autonomous driving. Yet, existing models trained on real-world datasets, which naturally exhibit long-tail distributions, tend to underperform on rare and safety-critical, vulnerable classes, such as pedestrians and cyclists. Existing studies on reweighting and resampling techniques struggle with the scarcity and limited diversity within tail classes. To address these limitations, we introduce LTDA-Drive, a novel LLM-guided data augmentation framework designed to synthesize diverse, high-quality long-tail samples. LTDA-Drive replaces head-class objects in driving scenes with tail-class objects through a three-stage process: (1) text-guided diffusion models remove head-class objects, (2) generative models insert instances of the tail classes, and (3) an LLM agent filters out low-quality synthesized images. Experiments conducted on the KITTI dataset show that LTDA-Drive significantly improves tail-class detection, achieving 34.75\% improvement for rare classes over counterpart methods. These results further highlight the effectiveness of LTDA-Drive in tackling long-tail challenges by generating high-quality and diverse data.
On the Foundation Model for Cardiac MRI Reconstruction
Nov 15, 2024



Abstract:In recent years, machine learning (ML) based reconstruction has been widely investigated and employed in cardiac magnetic resonance (CMR) imaging. ML-based reconstructions can deliver clinically acceptable image quality under substantially accelerated scans. ML-based reconstruction, however, also requires substantial data and computational time to train the neural network, which is often optimized for a fixed acceleration rate or image contrast. In practice, imaging parameters are often tuned to best suit the diagnosis, which may differ from the training data. This can result in degraded image quality, and multiple trained networks are needed to fulfill the clinical demands. In this study, we propose a foundation model that uses adaptive unrolling, channel-shifting, and Pattern and Contrast-Prompt-UNet (PCP-UNet) to tackle the problem. In particular, the undersampled data goes through a different number of unrolled iterations according to its acceleration rate. Channel-shifting improves reconstructed data quality. The PCP-UNet is equipped with an image contrast and sampling pattern prompt. In vivo CMR experiments were performed using mixed combinations of image contrasts, acceleration rates, and (under)sampling patterns. The proposed foundation model has significantly improved image quality for a wide range of CMR protocols and outperforms the conventional ML-based method.
I2I-Mamba: Multi-modal medical image synthesis via selective state space modeling
May 22, 2024



Abstract:In recent years, deep learning models comprising transformer components have pushed the performance envelope in medical image synthesis tasks. Contrary to convolutional neural networks (CNNs) that use static, local filters, transformers use self-attention mechanisms to permit adaptive, non-local filtering to sensitively capture long-range context. However, this sensitivity comes at the expense of substantial model complexity, which can compromise learning efficacy particularly on relatively modest-sized imaging datasets. Here, we propose a novel adversarial model for multi-modal medical image synthesis, I2I-Mamba, that leverages selective state space modeling (SSM) to efficiently capture long-range context while maintaining local precision. To do this, I2I-Mamba injects channel-mixed Mamba (cmMamba) blocks in the bottleneck of a convolutional backbone. In cmMamba blocks, SSM layers are used to learn context across the spatial dimension and channel-mixing layers are used to learn context across the channel dimension of feature maps. Comprehensive demonstrations are reported for imputing missing images in multi-contrast MRI and MRI-CT protocols. Our results indicate that I2I-Mamba offers superior performance against state-of-the-art CNN- and transformer-based methods in synthesizing target-modality images.
GRJointNET: Synergistic Completion and Part Segmentation on 3D Incomplete Point Clouds
Nov 23, 2023



Abstract:Segmentation of three-dimensional (3D) point clouds is an important task for autonomous systems. However, success of segmentation algorithms depends greatly on the quality of the underlying point clouds (resolution, completeness etc.). In particular, incomplete point clouds might reduce a downstream model's performance. GRNet is proposed as a novel and recent deep learning solution to complete point clouds, but it is not capable of part segmentation. On the other hand, our proposed solution, GRJointNet, is an architecture that can perform joint completion and segmentation on point clouds as a successor of GRNet. Features extracted for the two tasks are also utilized by each other to increase the overall performance. We evaluated our proposed network on the ShapeNet-Part dataset and compared its performance to GRNet. Our results demonstrate GRJointNet can outperform GRNet on point completion. It should also be noted that GRNet is not capable of segmentation while GRJointNet is. This study1, therefore, holds a promise to enhance practicality and utility of point clouds in 3D vision for autonomous systems.
DDM$^2$: Self-Supervised Diffusion MRI Denoising with Generative Diffusion Models
Feb 06, 2023Abstract:Magnetic resonance imaging (MRI) is a common and life-saving medical imaging technique. However, acquiring high signal-to-noise ratio MRI scans requires long scan times, resulting in increased costs and patient discomfort, and decreased throughput. Thus, there is great interest in denoising MRI scans, especially for the subtype of diffusion MRI scans that are severely SNR-limited. While most prior MRI denoising methods are supervised in nature, acquiring supervised training datasets for the multitude of anatomies, MRI scanners, and scan parameters proves impractical. Here, we propose Denoising Diffusion Models for Denoising Diffusion MRI (DDM$^2$), a self-supervised denoising method for MRI denoising using diffusion denoising generative models. Our three-stage framework integrates statistic-based denoising theory into diffusion models and performs denoising through conditional generation. During inference, we represent input noisy measurements as a sample from an intermediate posterior distribution within the diffusion Markov chain. We conduct experiments on 4 real-world in-vivo diffusion MRI datasets and show that our DDM$^2$ demonstrates superior denoising performances ascertained with clinically-relevant visual qualitative and quantitative metrics.
ResViT: Residual vision transformers for multi-modal medical image synthesis
Jun 30, 2021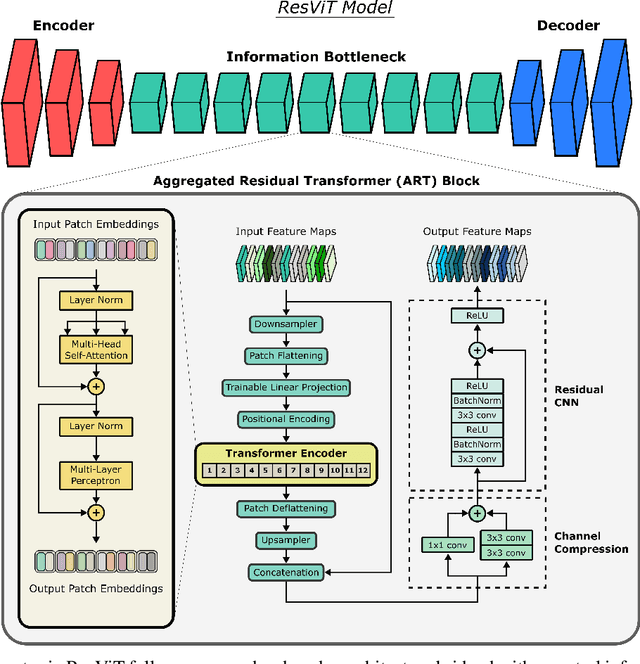
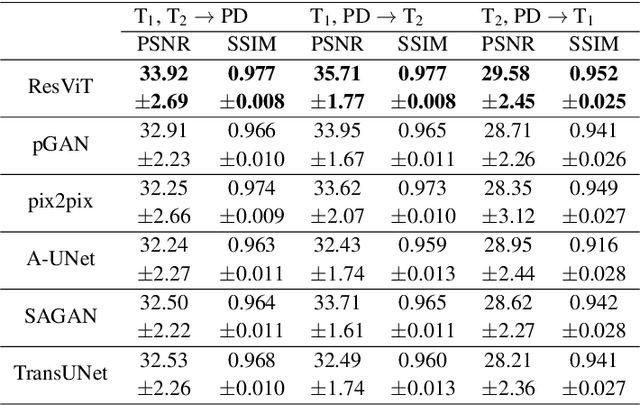
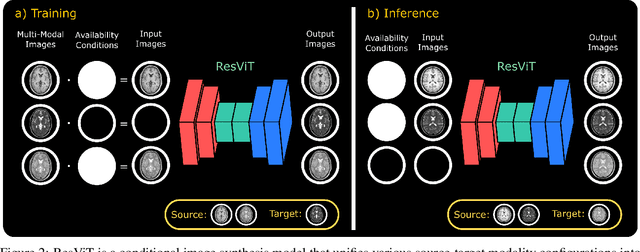
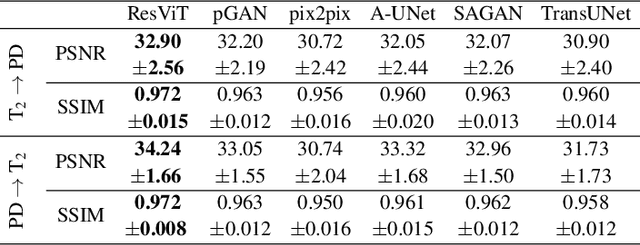
Abstract:Multi-modal imaging is a key healthcare technology in the diagnosis and management of disease, but it is often underutilized due to costs associated with multiple separate scans. This limitation yields the need for synthesis of unacquired modalities from the subset of available modalities. In recent years, generative adversarial network (GAN) models with superior depiction of structural details have been established as state-of-the-art in numerous medical image synthesis tasks. However, GANs are characteristically based on convolutional neural network (CNN) backbones that perform local processing with compact filters. This inductive bias, in turn, compromises learning of long-range spatial dependencies. While attention maps incorporated in GANs can multiplicatively modulate CNN features to emphasize critical image regions, their capture of global context is mostly implicit. Here, we propose a novel generative adversarial approach for medical image synthesis, ResViT, to combine local precision of convolution operators with contextual sensitivity of vision transformers. Based on an encoder-decoder architecture, ResViT employs a central bottleneck comprising novel aggregated residual transformer (ART) blocks that synergistically combine convolutional and transformer modules. Comprehensive demonstrations are performed for synthesizing missing sequences in multi-contrast MRI and CT images from MRI. Our results indicate the superiority of ResViT against competing methods in terms of qualitative observations and quantitative metrics.
Unsupervised MRI Reconstruction via Zero-Shot Learned Adversarial Transformers
May 21, 2021



Abstract:Supervised deep learning has swiftly become a workhorse for accelerated MRI in recent years, offering state-of-the-art performance in image reconstruction from undersampled acquisitions. Training deep supervised models requires large datasets of undersampled and fully-sampled acquisitions typically from a matching set of subjects. Given scarce access to large medical datasets, this limitation has sparked interest in unsupervised methods that reduce reliance on fully-sampled ground-truth data. A common framework is based on the deep image prior, where network-driven regularization is enforced directly during inference on undersampled acquisitions. Yet, canonical convolutional architectures are suboptimal in capturing long-range relationships, and randomly initialized networks may hamper convergence. To address these limitations, here we introduce a novel unsupervised MRI reconstruction method based on zero-Shot Learned Adversarial TransformERs (SLATER). SLATER embodies a deep adversarial network with cross-attention transformer blocks to map noise and latent variables onto MR images. This unconditional network learns a high-quality MRI prior in a self-supervised encoding task. A zero-shot reconstruction is performed on undersampled test data, where inference is performed by optimizing network parameters, latent and noise variables to ensure maximal consistency to multi-coil MRI data. Comprehensive experiments on brain MRI datasets clearly demonstrate the superior performance of SLATER against several state-of-the-art unsupervised methods.
A Few-Shot Learning Approach for Accelerated MRI via Fusion of Data-Driven and Subject-Driven Priors
Mar 13, 2021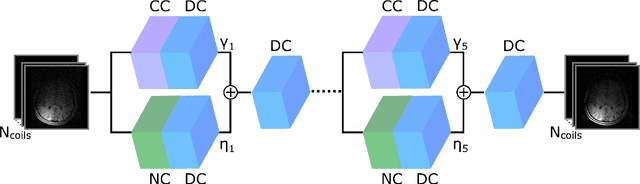
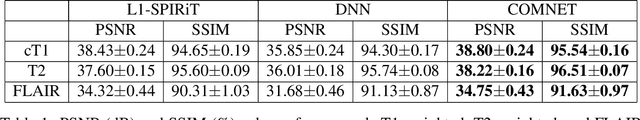
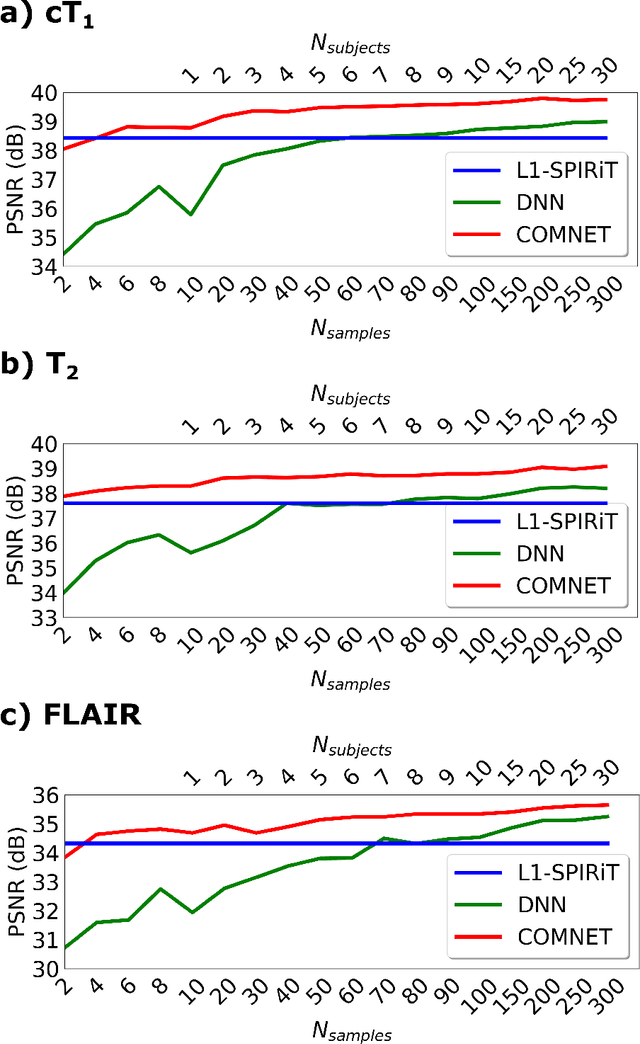
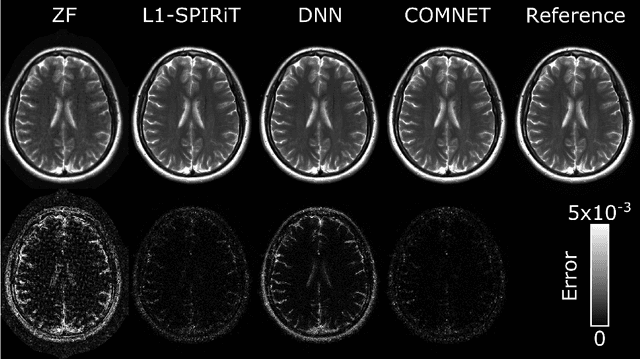
Abstract:Deep neural networks (DNNs) have recently found emerging use in accelerated MRI reconstruction. DNNs typically learn data-driven priors from large datasets constituting pairs of undersampled and fully-sampled acquisitions. Acquiring such large datasets, however, might be impractical. To mitigate this limitation, we propose a few-shot learning approach for accelerated MRI that merges subject-driven priors obtained via physical signal models with data-driven priors obtained from a few training samples. Demonstrations on brain MR images from the NYU fastMRI dataset indicate that the proposed approach requires just a few samples to outperform traditional parallel imaging and DNN algorithms.
Three Dimensional MR Image Synthesis with Progressive Generative Adversarial Networks
Dec 18, 2020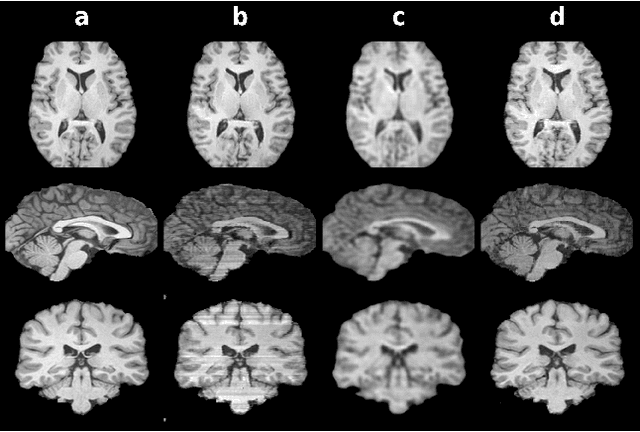
Abstract:Mainstream deep models for three-dimensional MRI synthesis are either cross-sectional or volumetric depending on the input. Cross-sectional models can decrease the model complexity, but they may lead to discontinuity artifacts. On the other hand, volumetric models can alleviate the discontinuity artifacts, but they might suffer from loss of spatial resolution due to increased model complexity coupled with scarce training data. To mitigate the limitations of both approaches, we propose a novel model that progressively recovers the target volume via simpler synthesis tasks across individual orientations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge