Salman UH Dar
Learning Fourier-Constrained Diffusion Bridges for MRI Reconstruction
Aug 04, 2023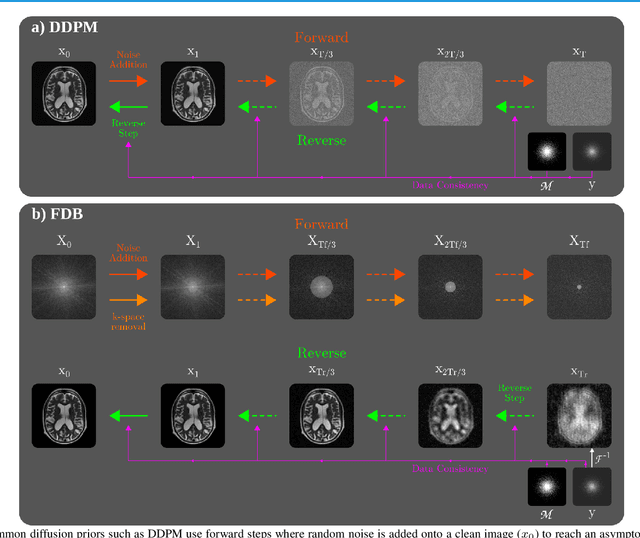
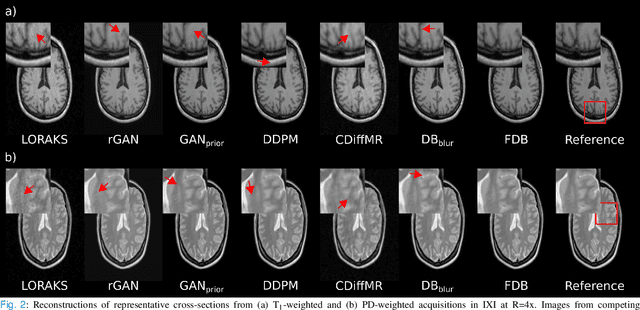
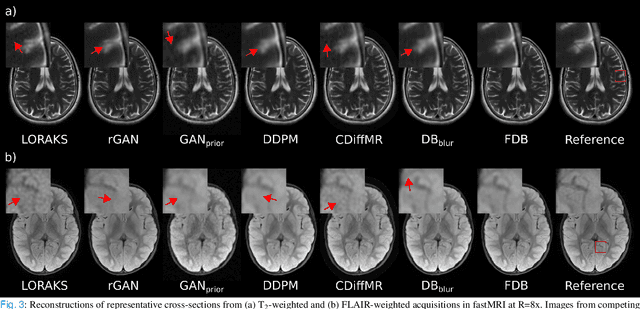
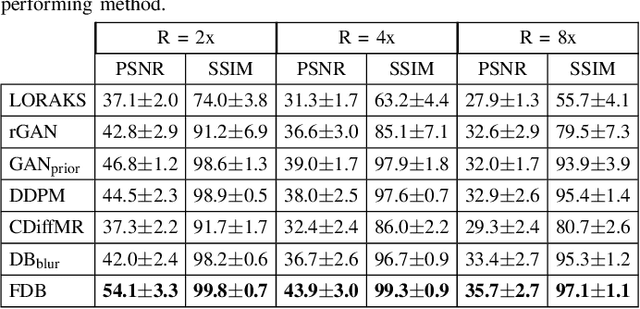
Abstract:Recent years have witnessed a surge in deep generative models for accelerated MRI reconstruction. Diffusion priors in particular have gained traction with their superior representational fidelity and diversity. Instead of the target transformation from undersampled to fully-sampled data, common diffusion priors are trained to learn a multi-step transformation from Gaussian noise onto fully-sampled data. During inference, data-fidelity projections are injected in between reverse diffusion steps to reach a compromise solution within the span of both the diffusion prior and the imaging operator. Unfortunately, suboptimal solutions can arise as the normality assumption of the diffusion prior causes divergence between learned and target transformations. To address this limitation, here we introduce the first diffusion bridge for accelerated MRI reconstruction. The proposed Fourier-constrained diffusion bridge (FDB) leverages a generalized process to transform between undersampled and fully-sampled data via random noise addition and random frequency removal as degradation operators. Unlike common diffusion priors that use an asymptotic endpoint based on Gaussian noise, FDB captures a transformation between finite endpoints where the initial endpoint is based on moderate degradation of fully-sampled data. Demonstrations on brain MRI indicate that FDB outperforms state-of-the-art reconstruction methods including conventional diffusion priors.
JointNET: A Deep Model for Predicting Active Sacroiliitis from Sacroiliac Joint Radiography
Jan 27, 2023Abstract:Purpose: To develop a deep learning model that predicts active inflammation from sacroiliac joint radiographs and to compare the success with radiologists. Materials and Methods: A total of 1,537 (augmented 1752) grade 0 SIJs of 768 patients were retrospectively analyzed. Gold-standard MRI exams showed active inflammation in 330 joints according to ASAS criteria. A convolutional neural network model (JointNET) was developed to detect MRI-based active inflammation labels solely based on radiographs. Two radiologists blindly evaluated the radiographs for comparison. Python, PyTorch, and SPSS were used for analyses. P<0.05 was considered statistically significant. Results: JointNET differentiated active inflammation from radiographs with a mean AUROC of 89.2 (95% CI:86.8%, 91.7%). The sensitivity was 69.0% (95% CI:65.3%, 72.7%) and specificity 90.4% (95% CI:87.8 % 92.9%). The mean accuracy was 90.2% (95% CI: 87.6%, 92.8%). The positive predictive value was 74.6% (95% CI: 72.5%, 76.7%) and negative predictive value was 87.9% (95% CI: 85.4%, 90.5%) when prevalence was considered 1%. Statistical analyses showed a significant difference between active inflammation and healthy groups (p<0.05). Radiologists accuracies were less than 65% to discriminate active inflammation from sacroiliac joint radiographs. Conclusion: JointNET successfully predicts active inflammation from sacroiliac joint radiographs, with superior performance to human observers.
Learning Deep MRI Reconstruction Models from Scratch in Low-Data Regimes
Jan 06, 2023Abstract:Magnetic resonance imaging (MRI) is an essential diagnostic tool that suffers from prolonged scan times. Reconstruction methods can alleviate this limitation by recovering clinically usable images from accelerated acquisitions. In particular, learning-based methods promise performance leaps by employing deep neural networks as data-driven priors. A powerful approach uses scan-specific (SS) priors that leverage information regarding the underlying physical signal model for reconstruction. SS priors are learned on each individual test scan without the need for a training dataset, albeit they suffer from computationally burdening inference with nonlinear networks. An alternative approach uses scan-general (SG) priors that instead leverage information regarding the latent features of MRI images for reconstruction. SG priors are frozen at test time for efficiency, albeit they require learning from a large training dataset. Here, we introduce a novel parallel-stream fusion model (PSFNet) that synergistically fuses SS and SG priors for performant MRI reconstruction in low-data regimes, while maintaining competitive inference times to SG methods. PSFNet implements its SG prior based on a nonlinear network, yet it forms its SS prior based on a linear network to maintain efficiency. A pervasive framework for combining multiple priors in MRI reconstruction is algorithmic unrolling that uses serially alternated projections, causing error propagation under low-data regimes. To alleviate error propagation, PSFNet combines its SS and SG priors via a novel parallel-stream architecture with learnable fusion parameters. Demonstrations are performed on multi-coil brain MRI for varying amounts of training data. PSFNet outperforms SG methods in low-data regimes, and surpasses SS methods with few tens of training samples.
Unsupervised Medical Image Translation with Adversarial Diffusion Models
Jul 17, 2022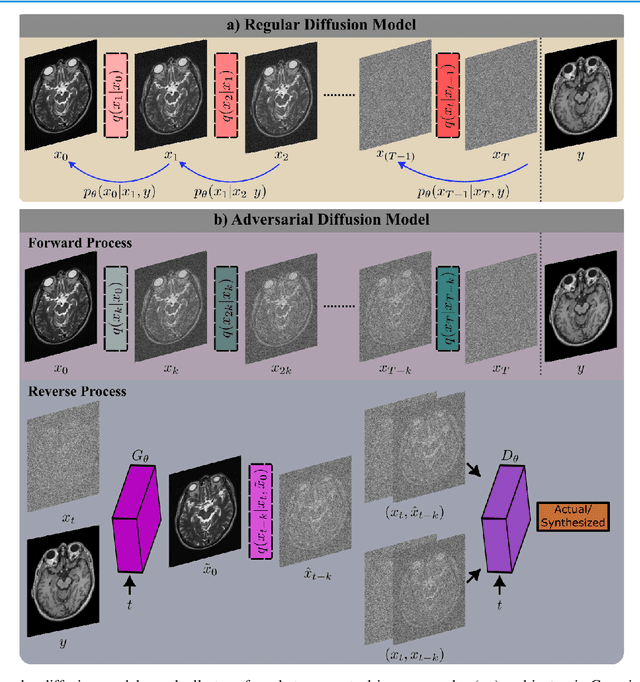
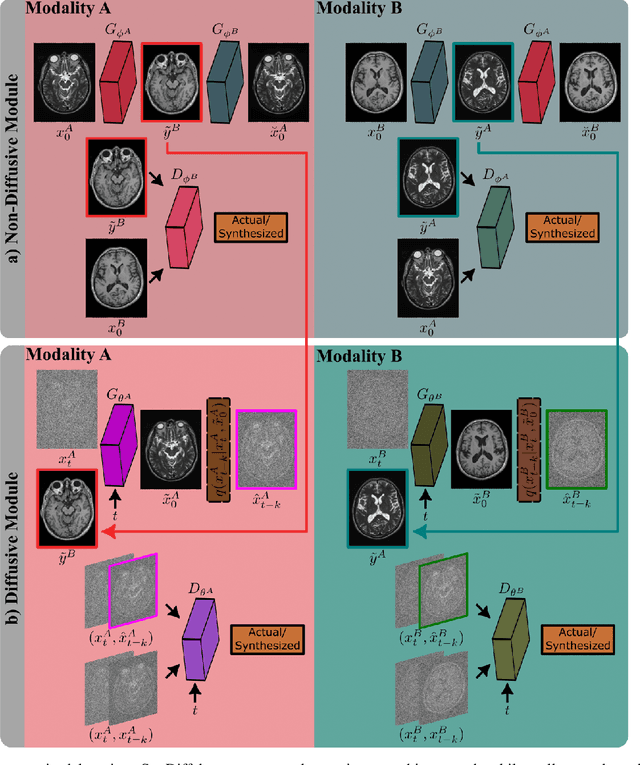
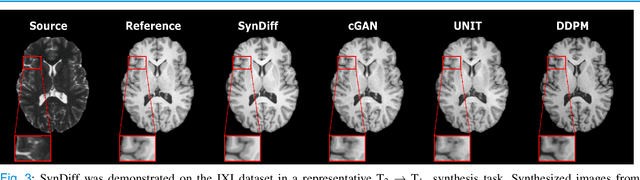
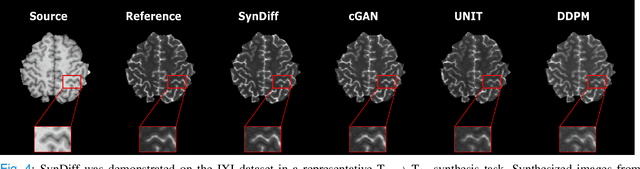
Abstract:Imputation of missing images via source-to-target modality translation can facilitate downstream tasks in medical imaging. A pervasive approach for synthesizing target images involves one-shot mapping through generative adversarial networks (GAN). Yet, GAN models that implicitly characterize the image distribution can suffer from limited sample fidelity and diversity. Here, we propose a novel method based on adversarial diffusion modeling, SynDiff, for improved reliability in medical image synthesis. To capture a direct correlate of the image distribution, SynDiff leverages a conditional diffusion process to progressively map noise and source images onto the target image. For fast and accurate image sampling during inference, large diffusion steps are coupled with adversarial projections in the reverse diffusion direction. To enable training on unpaired datasets, a cycle-consistent architecture is devised with two coupled diffusion processes to synthesize the target given source and the source given target. Extensive assessments are reported on the utility of SynDiff against competing GAN and diffusion models in multi-contrast MRI and MRI-CT translation. Our demonstrations indicate that SynDiff offers superior performance against competing baselines both qualitatively and quantitatively.
One Model to Unite Them All: Personalized Federated Learning of Multi-Contrast MRI Synthesis
Jul 13, 2022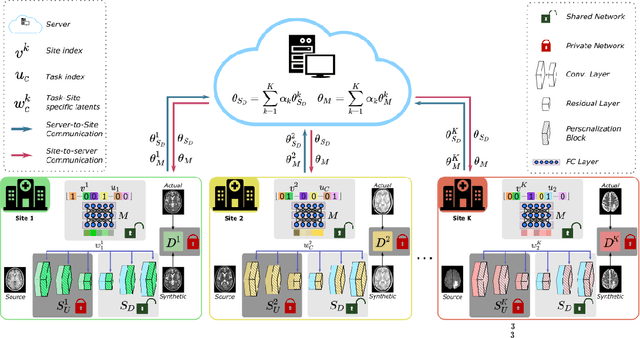
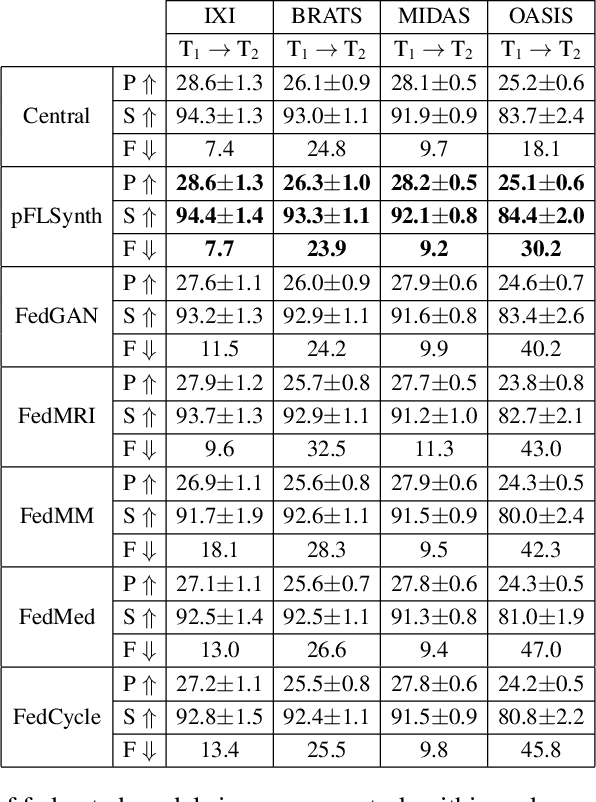
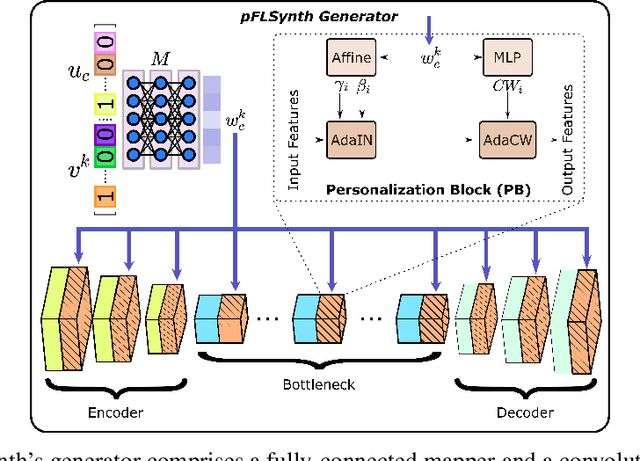
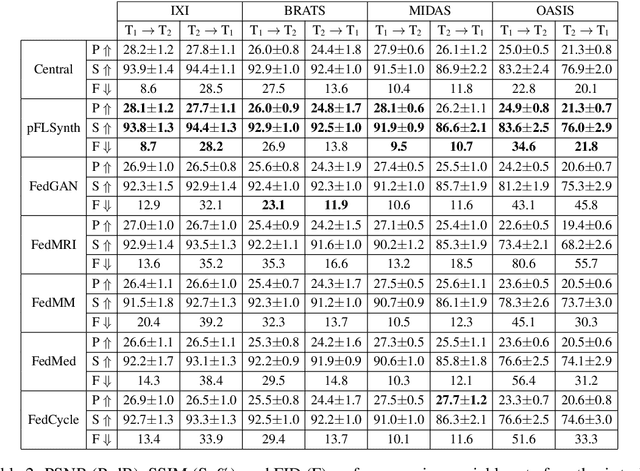
Abstract:Learning-based MRI translation involves a synthesis model that maps a source-contrast onto a target-contrast image. Multi-institutional collaborations are key to training synthesis models across broad datasets, yet centralized training involves privacy risks. Federated learning (FL) is a collaboration framework that instead adopts decentralized training to avoid sharing imaging data and mitigate privacy concerns. However, FL-trained models can be impaired by the inherent heterogeneity in the distribution of imaging data. On the one hand, implicit shifts in image distribution are evident across sites, even for a common translation task with fixed source-target configuration. Conversely, explicit shifts arise within and across sites when diverse translation tasks with varying source-target configurations are prescribed. To improve reliability against domain shifts, here we introduce the first personalized FL method for MRI Synthesis (pFLSynth). pFLSynth is based on an adversarial model equipped with a mapper that produces latents specific to individual sites and source-target contrasts. It leverages novel personalization blocks that adaptively tune the statistics and weighting of feature maps across the generator based on these latents. To further promote site-specificity, partial model aggregation is employed over downstream layers of the generator while upstream layers are retained locally. As such, pFLSynth enables training of a unified synthesis model that can reliably generalize across multiple sites and translation tasks. Comprehensive experiments on multi-site datasets clearly demonstrate the enhanced performance of pFLSynth against prior federated methods in multi-contrast MRI synthesis.
Adaptive Diffusion Priors for Accelerated MRI Reconstruction
Jul 12, 2022
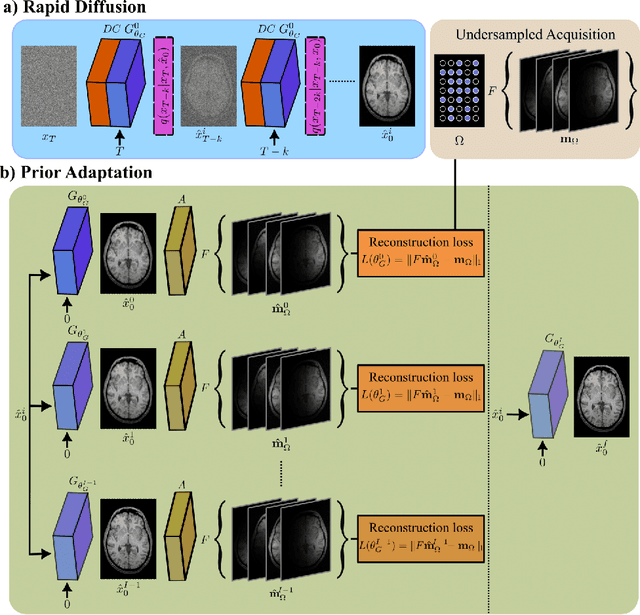
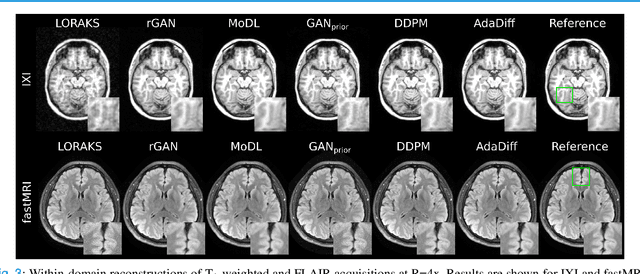
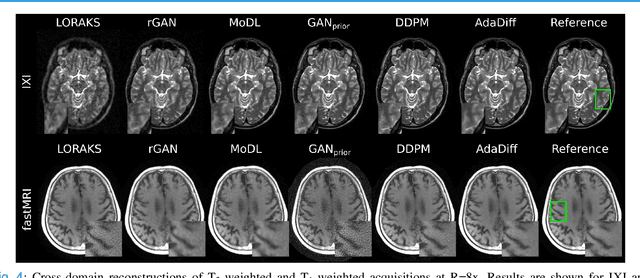
Abstract:Deep MRI reconstruction is commonly performed with conditional models that map undersampled data as input onto fully-sampled data as output. Conditional models perform de-aliasing under knowledge of the accelerated imaging operator, so they poorly generalize under domain shifts in the operator. Unconditional models are a powerful alternative that instead learn generative image priors to improve reliability against domain shifts. Recent diffusion models are particularly promising given their high representational diversity and sample quality. Nevertheless, projections through a static image prior can lead to suboptimal performance. Here we propose a novel MRI reconstruction, AdaDiff, based on an adaptive diffusion prior. To enable efficient image sampling, an adversarial mapper is introduced that enables use of large diffusion steps. A two-phase reconstruction is performed with the trained prior: a rapid-diffusion phase that produces an initial reconstruction, and an adaptation phase where the diffusion prior is updated to minimize reconstruction loss on acquired k-space data. Demonstrations on multi-contrast brain MRI clearly indicate that AdaDiff achieves superior performance to competing models in cross-domain tasks, and superior or on par performance in within-domain tasks.
Federated Learning of Generative Image Priors for MRI Reconstruction
Feb 08, 2022



Abstract:Multi-institutional efforts can facilitate training of deep MRI reconstruction models, albeit privacy risks arise during cross-site sharing of imaging data. Federated learning (FL) has recently been introduced to address privacy concerns by enabling distributed training without transfer of imaging data. Existing FL methods for MRI reconstruction employ conditional models to map from undersampled to fully-sampled acquisitions via explicit knowledge of the imaging operator. Since conditional models generalize poorly across different acceleration rates or sampling densities, imaging operators must be fixed between training and testing, and they are typically matched across sites. To improve generalization and flexibility in multi-institutional collaborations, here we introduce a novel method for MRI reconstruction based on Federated learning of Generative IMage Priors (FedGIMP). FedGIMP leverages a two-stage approach: cross-site learning of a generative MRI prior, and subject-specific injection of the imaging operator. The global MRI prior is learned via an unconditional adversarial model that synthesizes high-quality MR images based on latent variables. Specificity in the prior is preserved via a mapper subnetwork that produces site-specific latents. During inference, the prior is combined with subject-specific imaging operators to enable reconstruction, and further adapted to individual test samples by minimizing data-consistency loss. Comprehensive experiments on multi-institutional datasets clearly demonstrate enhanced generalization performance of FedGIMP against site-specific and federated methods based on conditional models, as well as traditional reconstruction methods.
Unsupervised MRI Reconstruction via Zero-Shot Learned Adversarial Transformers
May 21, 2021



Abstract:Supervised deep learning has swiftly become a workhorse for accelerated MRI in recent years, offering state-of-the-art performance in image reconstruction from undersampled acquisitions. Training deep supervised models requires large datasets of undersampled and fully-sampled acquisitions typically from a matching set of subjects. Given scarce access to large medical datasets, this limitation has sparked interest in unsupervised methods that reduce reliance on fully-sampled ground-truth data. A common framework is based on the deep image prior, where network-driven regularization is enforced directly during inference on undersampled acquisitions. Yet, canonical convolutional architectures are suboptimal in capturing long-range relationships, and randomly initialized networks may hamper convergence. To address these limitations, here we introduce a novel unsupervised MRI reconstruction method based on zero-Shot Learned Adversarial TransformERs (SLATER). SLATER embodies a deep adversarial network with cross-attention transformer blocks to map noise and latent variables onto MR images. This unconditional network learns a high-quality MRI prior in a self-supervised encoding task. A zero-shot reconstruction is performed on undersampled test data, where inference is performed by optimizing network parameters, latent and noise variables to ensure maximal consistency to multi-coil MRI data. Comprehensive experiments on brain MRI datasets clearly demonstrate the superior performance of SLATER against several state-of-the-art unsupervised methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge