Hasan A. Bedel
Learning Fourier-Constrained Diffusion Bridges for MRI Reconstruction
Aug 04, 2023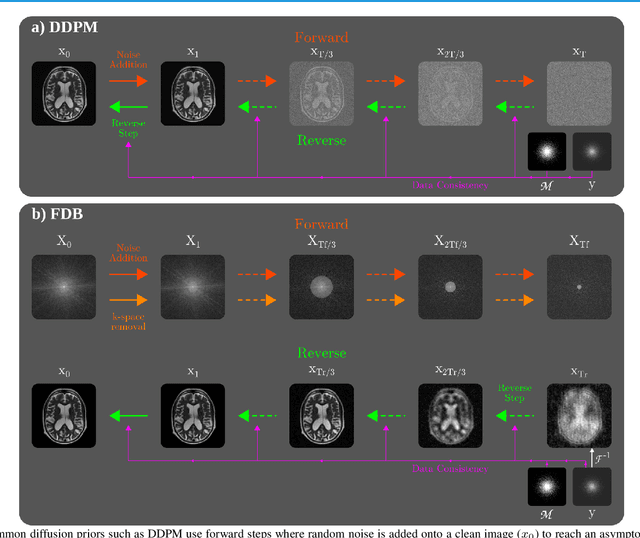
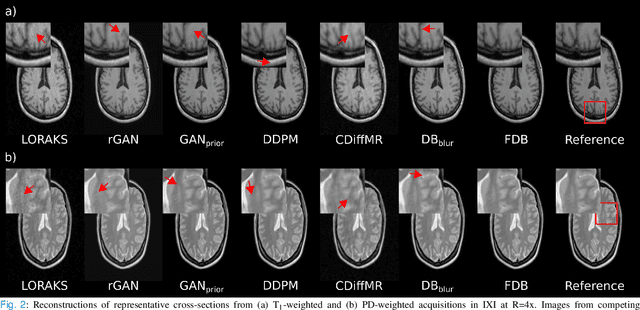
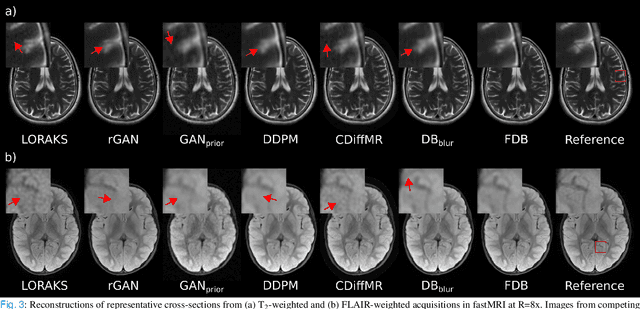
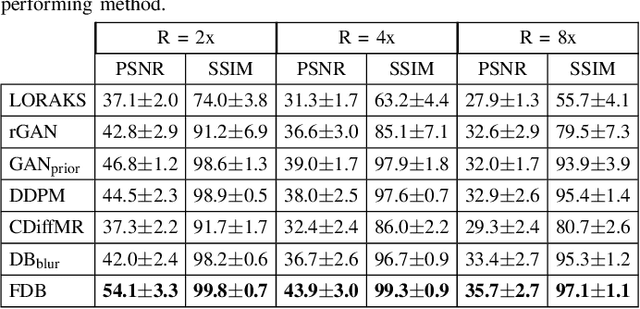
Abstract:Recent years have witnessed a surge in deep generative models for accelerated MRI reconstruction. Diffusion priors in particular have gained traction with their superior representational fidelity and diversity. Instead of the target transformation from undersampled to fully-sampled data, common diffusion priors are trained to learn a multi-step transformation from Gaussian noise onto fully-sampled data. During inference, data-fidelity projections are injected in between reverse diffusion steps to reach a compromise solution within the span of both the diffusion prior and the imaging operator. Unfortunately, suboptimal solutions can arise as the normality assumption of the diffusion prior causes divergence between learned and target transformations. To address this limitation, here we introduce the first diffusion bridge for accelerated MRI reconstruction. The proposed Fourier-constrained diffusion bridge (FDB) leverages a generalized process to transform between undersampled and fully-sampled data via random noise addition and random frequency removal as degradation operators. Unlike common diffusion priors that use an asymptotic endpoint based on Gaussian noise, FDB captures a transformation between finite endpoints where the initial endpoint is based on moderate degradation of fully-sampled data. Demonstrations on brain MRI indicate that FDB outperforms state-of-the-art reconstruction methods including conventional diffusion priors.
A plug-in graph neural network to boost temporal sensitivity in fMRI analysis
Jan 01, 2023Abstract:Learning-based methods have recently enabled performance leaps in analysis of high-dimensional functional MRI (fMRI) time series. Deep learning models that receive as input functional connectivity (FC) features among brain regions have been commonly adopted in the literature. However, many models focus on temporally static FC features across a scan, reducing sensitivity to dynamic features of brain activity. Here, we describe a plug-in graph neural network that can be flexibly integrated into a main learning-based fMRI model to boost its temporal sensitivity. Receiving brain regions as nodes and blood-oxygen-level-dependent (BOLD) signals as node inputs, the proposed GraphCorr method leverages a node embedder module based on a transformer encoder to capture temporally-windowed latent representations of BOLD signals. GraphCorr also leverages a lag filter module to account for delayed interactions across nodes by computing cross-correlation of windowed BOLD signals across a range of time lags. Information captured by the two modules is fused via a message passing algorithm executed on the graph, and enhanced node features are then computed at the output. These enhanced features are used to drive a subsequent learning-based model to analyze fMRI time series with elevated sensitivity. Comprehensive demonstrations on two public datasets indicate improved classification performance and interpretability for several state-of-the-art graphical and convolutional methods that employ GraphCorr-derived feature representations of fMRI time series as their input.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge