Şaban Öztürk
HydraViT: Adaptive Multi-Branch Transformer for Multi-Label Disease Classification from Chest X-ray Images
Oct 09, 2023Abstract:Chest X-ray is an essential diagnostic tool in the identification of chest diseases given its high sensitivity to pathological abnormalities in the lungs. However, image-driven diagnosis is still challenging due to heterogeneity in size and location of pathology, as well as visual similarities and co-occurrence of separate pathology. Since disease-related regions often occupy a relatively small portion of diagnostic images, classification models based on traditional convolutional neural networks (CNNs) are adversely affected given their locality bias. While CNNs were previously augmented with attention maps or spatial masks to guide focus on potentially critical regions, learning localization guidance under heterogeneity in the spatial distribution of pathology is challenging. To improve multi-label classification performance, here we propose a novel method, HydraViT, that synergistically combines a transformer backbone with a multi-branch output module with learned weighting. The transformer backbone enhances sensitivity to long-range context in X-ray images, while using the self-attention mechanism to adaptively focus on task-critical regions. The multi-branch output module dedicates an independent branch to each disease label to attain robust learning across separate disease classes, along with an aggregated branch across labels to maintain sensitivity to co-occurrence relationships among pathology. Experiments demonstrate that, on average, HydraViT outperforms competing attention-guided methods by 1.2%, region-guided methods by 1.4%, and semantic-guided methods by 1.0% in multi-label classification performance.
Learning Deep MRI Reconstruction Models from Scratch in Low-Data Regimes
Jan 06, 2023Abstract:Magnetic resonance imaging (MRI) is an essential diagnostic tool that suffers from prolonged scan times. Reconstruction methods can alleviate this limitation by recovering clinically usable images from accelerated acquisitions. In particular, learning-based methods promise performance leaps by employing deep neural networks as data-driven priors. A powerful approach uses scan-specific (SS) priors that leverage information regarding the underlying physical signal model for reconstruction. SS priors are learned on each individual test scan without the need for a training dataset, albeit they suffer from computationally burdening inference with nonlinear networks. An alternative approach uses scan-general (SG) priors that instead leverage information regarding the latent features of MRI images for reconstruction. SG priors are frozen at test time for efficiency, albeit they require learning from a large training dataset. Here, we introduce a novel parallel-stream fusion model (PSFNet) that synergistically fuses SS and SG priors for performant MRI reconstruction in low-data regimes, while maintaining competitive inference times to SG methods. PSFNet implements its SG prior based on a nonlinear network, yet it forms its SS prior based on a linear network to maintain efficiency. A pervasive framework for combining multiple priors in MRI reconstruction is algorithmic unrolling that uses serially alternated projections, causing error propagation under low-data regimes. To alleviate error propagation, PSFNet combines its SS and SG priors via a novel parallel-stream architecture with learnable fusion parameters. Demonstrations are performed on multi-coil brain MRI for varying amounts of training data. PSFNet outperforms SG methods in low-data regimes, and surpasses SS methods with few tens of training samples.
A plug-in graph neural network to boost temporal sensitivity in fMRI analysis
Jan 01, 2023Abstract:Learning-based methods have recently enabled performance leaps in analysis of high-dimensional functional MRI (fMRI) time series. Deep learning models that receive as input functional connectivity (FC) features among brain regions have been commonly adopted in the literature. However, many models focus on temporally static FC features across a scan, reducing sensitivity to dynamic features of brain activity. Here, we describe a plug-in graph neural network that can be flexibly integrated into a main learning-based fMRI model to boost its temporal sensitivity. Receiving brain regions as nodes and blood-oxygen-level-dependent (BOLD) signals as node inputs, the proposed GraphCorr method leverages a node embedder module based on a transformer encoder to capture temporally-windowed latent representations of BOLD signals. GraphCorr also leverages a lag filter module to account for delayed interactions across nodes by computing cross-correlation of windowed BOLD signals across a range of time lags. Information captured by the two modules is fused via a message passing algorithm executed on the graph, and enhanced node features are then computed at the output. These enhanced features are used to drive a subsequent learning-based model to analyze fMRI time series with elevated sensitivity. Comprehensive demonstrations on two public datasets indicate improved classification performance and interpretability for several state-of-the-art graphical and convolutional methods that employ GraphCorr-derived feature representations of fMRI time series as their input.
Content-Based Medical Image Retrieval with Opponent Class Adaptive Margin Loss
Nov 22, 2022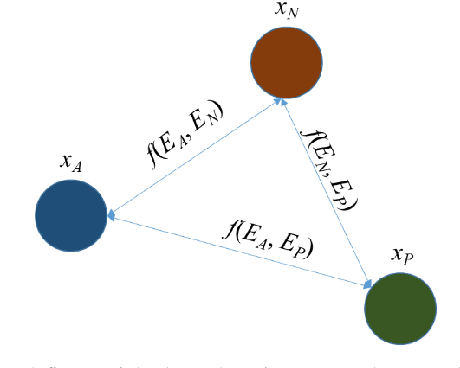
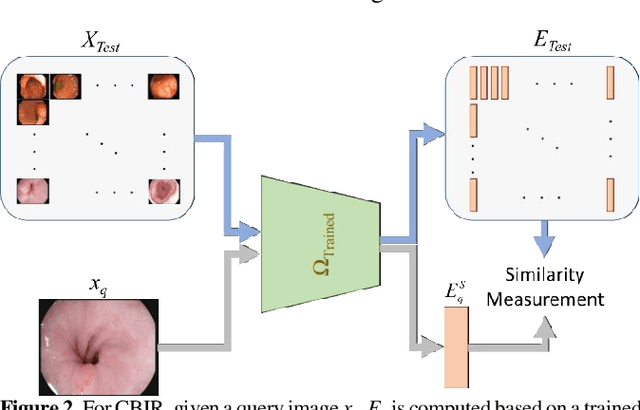
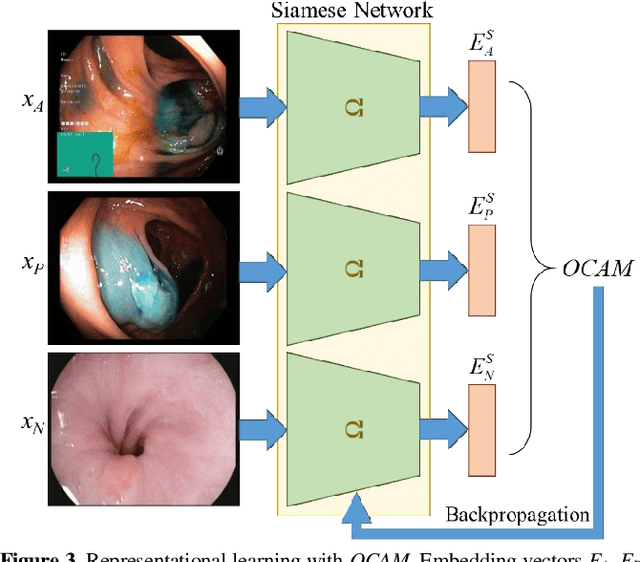
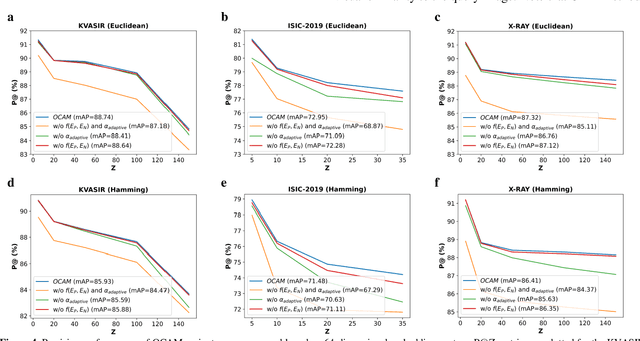
Abstract:Broadspread use of medical imaging devices with digital storage has paved the way for curation of substantial data repositories. Fast access to image samples with similar appearance to suspected cases can help establish a consulting system for healthcare professionals, and improve diagnostic procedures while minimizing processing delays. However, manual querying of large data repositories is labor intensive. Content-based image retrieval (CBIR) offers an automated solution based on dense embedding vectors that represent image features to allow quantitative similarity assessments. Triplet learning has emerged as a powerful approach to recover embeddings in CBIR, albeit traditional loss functions ignore the dynamic relationship between opponent image classes. Here, we introduce a triplet-learning method for automated querying of medical image repositories based on a novel Opponent Class Adaptive Margin (OCAM) loss. OCAM uses a variable margin value that is updated continually during the course of training to maintain optimally discriminative representations. CBIR performance of OCAM is compared against state-of-the-art loss functions for representational learning on three public databases (gastrointestinal disease, skin lesion, lung disease). Comprehensive experiments in each application domain demonstrate the superior performance of OCAM against baselines.
Adaptive Diffusion Priors for Accelerated MRI Reconstruction
Jul 12, 2022
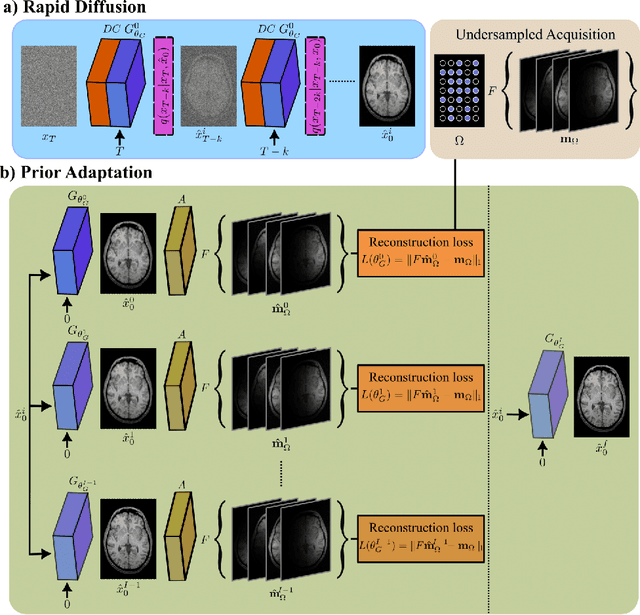
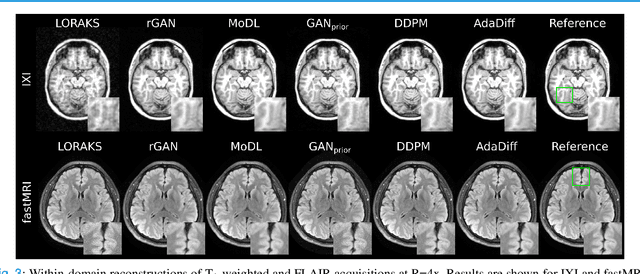
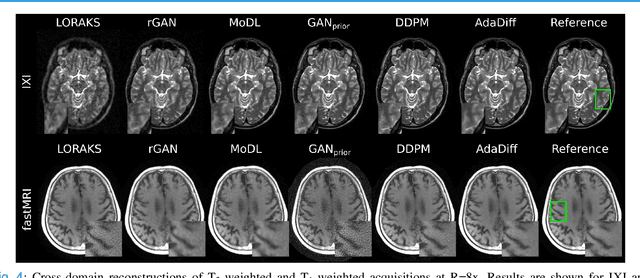
Abstract:Deep MRI reconstruction is commonly performed with conditional models that map undersampled data as input onto fully-sampled data as output. Conditional models perform de-aliasing under knowledge of the accelerated imaging operator, so they poorly generalize under domain shifts in the operator. Unconditional models are a powerful alternative that instead learn generative image priors to improve reliability against domain shifts. Recent diffusion models are particularly promising given their high representational diversity and sample quality. Nevertheless, projections through a static image prior can lead to suboptimal performance. Here we propose a novel MRI reconstruction, AdaDiff, based on an adaptive diffusion prior. To enable efficient image sampling, an adversarial mapper is introduced that enables use of large diffusion steps. A two-phase reconstruction is performed with the trained prior: a rapid-diffusion phase that produces an initial reconstruction, and an adaptation phase where the diffusion prior is updated to minimize reconstruction loss on acquired k-space data. Demonstrations on multi-contrast brain MRI clearly indicate that AdaDiff achieves superior performance to competing models in cross-domain tasks, and superior or on par performance in within-domain tasks.
Classification of COVID-19 in Chest CT Images using Convolutional Support Vector Machines
Nov 11, 2020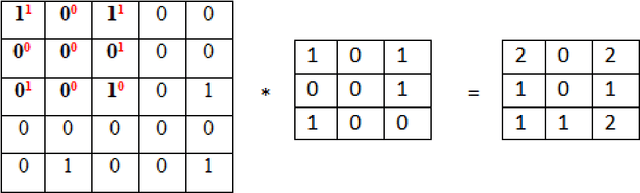
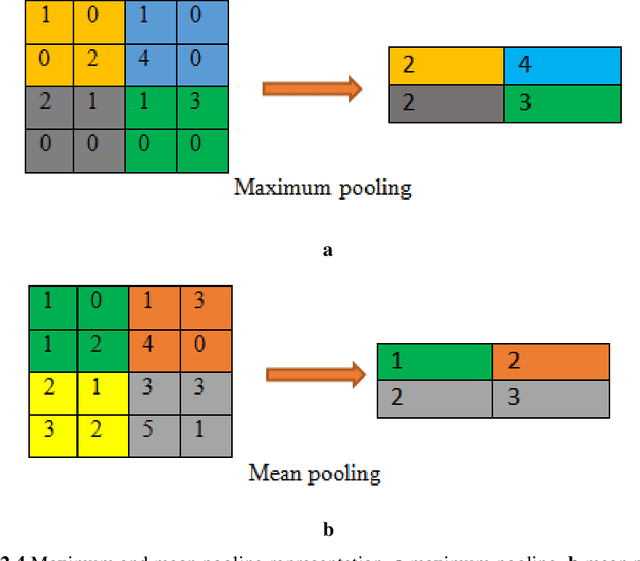
Abstract:Purpose: Coronavirus 2019 (COVID-19), which emerged in Wuhan, China and affected the whole world, has cost the lives of thousands of people. Manual diagnosis is inefficient due to the rapid spread of this virus. For this reason, automatic COVID-19 detection studies are carried out with the support of artificial intelligence algorithms. Methods: In this study, a deep learning model that detects COVID-19 cases with high performance is presented. The proposed method is defined as Convolutional Support Vector Machine (CSVM) and can automatically classify Computed Tomography (CT) images. Unlike the pre-trained Convolutional Neural Networks (CNN) trained with the transfer learning method, the CSVM model is trained as a scratch. To evaluate the performance of the CSVM method, the dataset is divided into two parts as training (%75) and testing (%25). The CSVM model consists of blocks containing three different numbers of SVM kernels. Results: When the performance of pre-trained CNN networks and CSVM models is assessed, CSVM (7x7, 3x3, 1x1) model shows the highest performance with 94.03% ACC, 96.09% SEN, 92.01% SPE, 92.19% PRE, 94.10% F1-Score, 88.15% MCC and 88.07% Kappa metric values. Conclusion: The proposed method is more effective than other methods. It has proven in experiments performed to be an inspiration for combating COVID and for future studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge