Tolga Cukur
Self-Supervised MRI Reconstruction with Unrolled Diffusion Models
Jun 29, 2023Abstract:Magnetic Resonance Imaging (MRI) produces excellent soft tissue contrast, albeit it is an inherently slow imaging modality. Promising deep learning methods have recently been proposed to reconstruct accelerated MRI scans. However, existing methods still suffer from various limitations regarding image fidelity, contextual sensitivity, and reliance on fully-sampled acquisitions for model training. To comprehensively address these limitations, we propose a novel self-supervised deep reconstruction model, named Self-Supervised Diffusion Reconstruction (SSDiffRecon). SSDiffRecon expresses a conditional diffusion process as an unrolled architecture that interleaves cross-attention transformers for reverse diffusion steps with data-consistency blocks for physics-driven processing. Unlike recent diffusion methods for MRI reconstruction, a self-supervision strategy is adopted to train SSDiffRecon using only undersampled k-space data. Comprehensive experiments on public brain MR datasets demonstrates the superiority of SSDiffRecon against state-of-the-art supervised, and self-supervised baselines in terms of reconstruction speed and quality. Implementation will be available at https://github.com/yilmazkorkmaz1/SSDiffRecon.
JointNET: A Deep Model for Predicting Active Sacroiliitis from Sacroiliac Joint Radiography
Jan 27, 2023Abstract:Purpose: To develop a deep learning model that predicts active inflammation from sacroiliac joint radiographs and to compare the success with radiologists. Materials and Methods: A total of 1,537 (augmented 1752) grade 0 SIJs of 768 patients were retrospectively analyzed. Gold-standard MRI exams showed active inflammation in 330 joints according to ASAS criteria. A convolutional neural network model (JointNET) was developed to detect MRI-based active inflammation labels solely based on radiographs. Two radiologists blindly evaluated the radiographs for comparison. Python, PyTorch, and SPSS were used for analyses. P<0.05 was considered statistically significant. Results: JointNET differentiated active inflammation from radiographs with a mean AUROC of 89.2 (95% CI:86.8%, 91.7%). The sensitivity was 69.0% (95% CI:65.3%, 72.7%) and specificity 90.4% (95% CI:87.8 % 92.9%). The mean accuracy was 90.2% (95% CI: 87.6%, 92.8%). The positive predictive value was 74.6% (95% CI: 72.5%, 76.7%) and negative predictive value was 87.9% (95% CI: 85.4%, 90.5%) when prevalence was considered 1%. Statistical analyses showed a significant difference between active inflammation and healthy groups (p<0.05). Radiologists accuracies were less than 65% to discriminate active inflammation from sacroiliac joint radiographs. Conclusion: JointNET successfully predicts active inflammation from sacroiliac joint radiographs, with superior performance to human observers.
Content-Based Medical Image Retrieval with Opponent Class Adaptive Margin Loss
Nov 22, 2022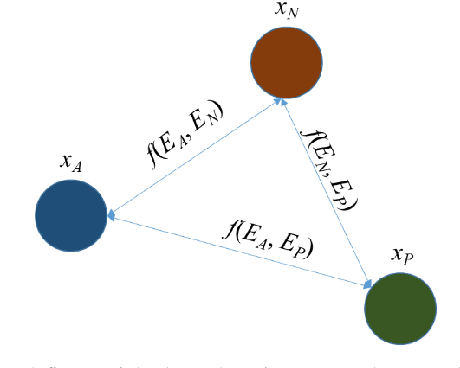
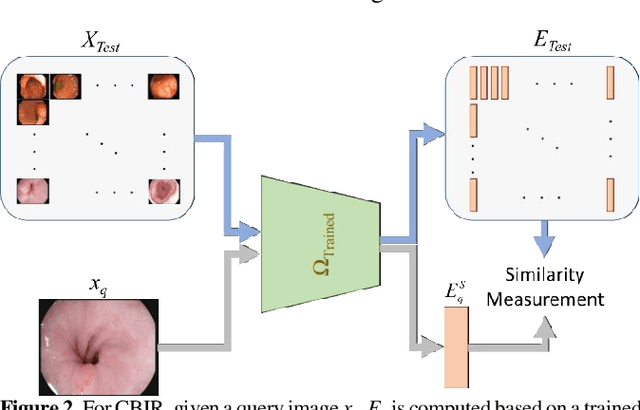
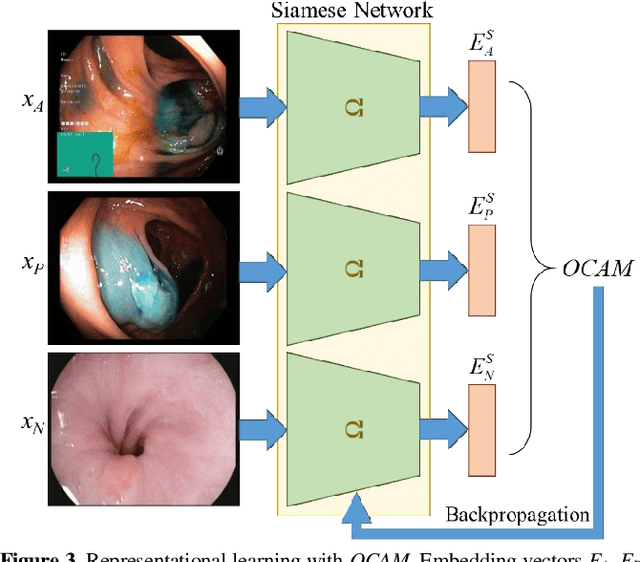
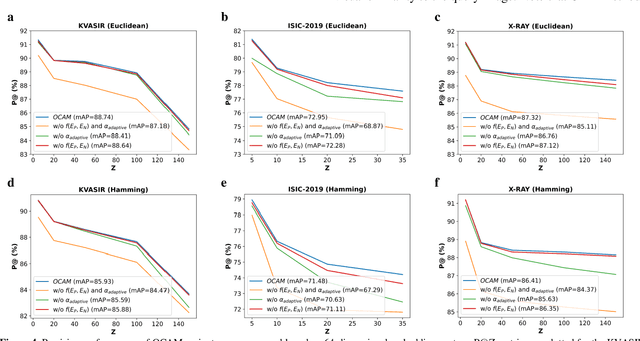
Abstract:Broadspread use of medical imaging devices with digital storage has paved the way for curation of substantial data repositories. Fast access to image samples with similar appearance to suspected cases can help establish a consulting system for healthcare professionals, and improve diagnostic procedures while minimizing processing delays. However, manual querying of large data repositories is labor intensive. Content-based image retrieval (CBIR) offers an automated solution based on dense embedding vectors that represent image features to allow quantitative similarity assessments. Triplet learning has emerged as a powerful approach to recover embeddings in CBIR, albeit traditional loss functions ignore the dynamic relationship between opponent image classes. Here, we introduce a triplet-learning method for automated querying of medical image repositories based on a novel Opponent Class Adaptive Margin (OCAM) loss. OCAM uses a variable margin value that is updated continually during the course of training to maintain optimally discriminative representations. CBIR performance of OCAM is compared against state-of-the-art loss functions for representational learning on three public databases (gastrointestinal disease, skin lesion, lung disease). Comprehensive experiments in each application domain demonstrate the superior performance of OCAM against baselines.
COVID-19 Detection from Respiratory Sounds with Hierarchical Spectrogram Transformers
Jul 19, 2022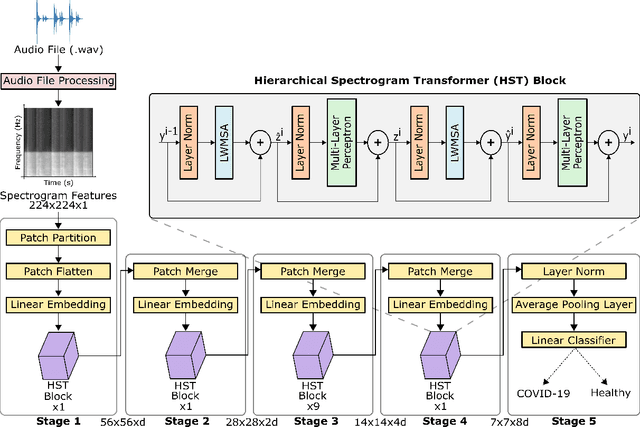
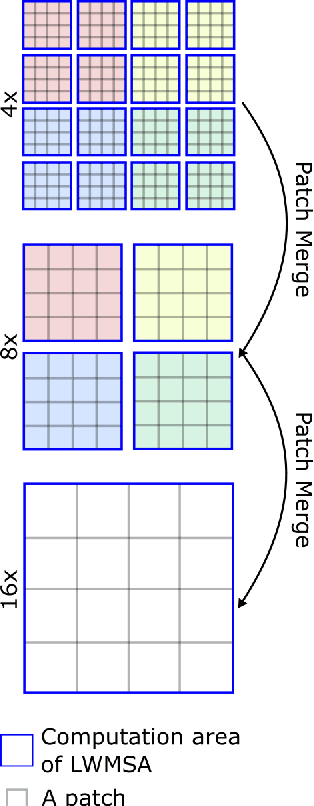
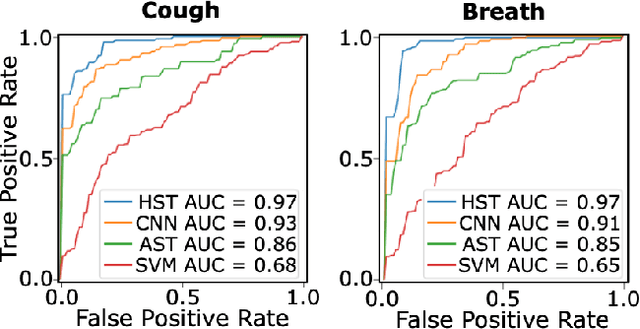
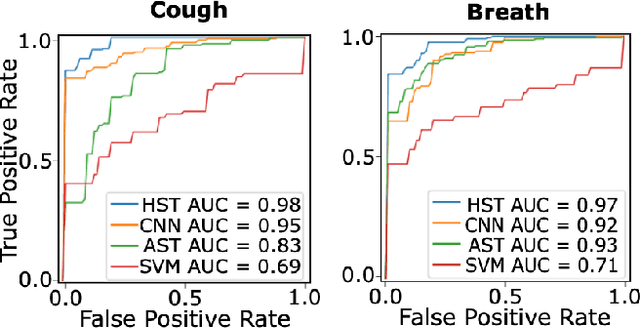
Abstract:Monitoring of prevalent airborne diseases such as COVID-19 characteristically involve respiratory assessments. While auscultation is a mainstream method for symptomatic monitoring, its diagnostic utility is hampered by the need for dedicated hospital visits. Continual remote monitoring based on recordings of respiratory sounds on portable devices is a promising alternative, which can assist in screening of COVID-19. In this study, we introduce a novel deep learning approach to distinguish patients with COVID-19 from healthy controls given audio recordings of cough or breathing sounds. The proposed approach leverages a novel hierarchical spectrogram transformer (HST) on spectrogram representations of respiratory sounds. HST embodies self-attention mechanisms over local windows in spectrograms, and window size is progressively grown over model stages to capture local to global context. HST is compared against state-of-the-art conventional and deep-learning baselines. Comprehensive demonstrations on a multi-national dataset indicate that HST outperforms competing methods, achieving over 97% area under the receiver operating characteristic curve (AUC) in detecting COVID-19 cases.
Deep Clustering via Center-Oriented Margin Free-Triplet Loss for Skin Lesion Detection in Highly Imbalanced Datasets
Apr 03, 2022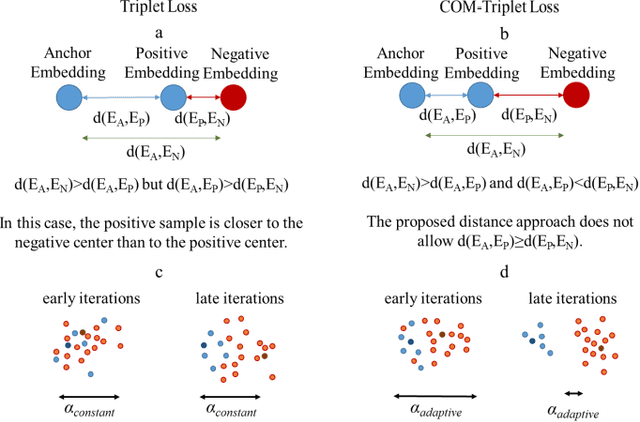
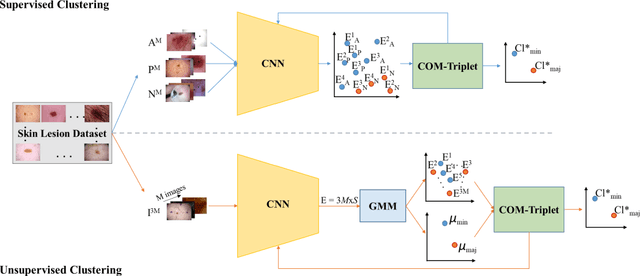
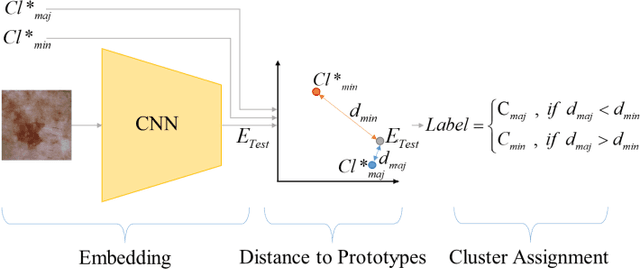
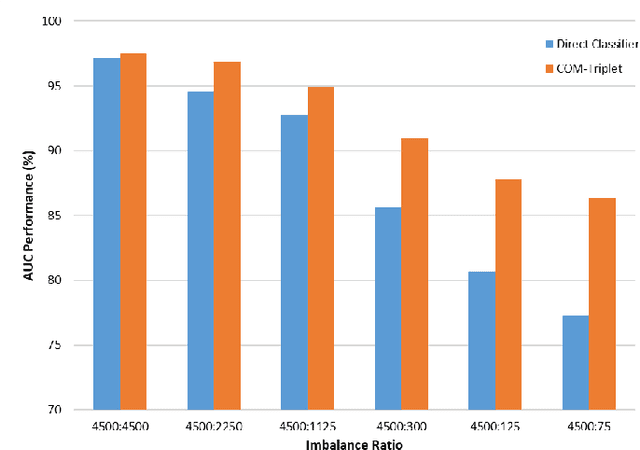
Abstract:Melanoma is a fatal skin cancer that is curable and has dramatically increasing survival rate when diagnosed at early stages. Learning-based methods hold significant promise for the detection of melanoma from dermoscopic images. However, since melanoma is a rare disease, existing databases of skin lesions predominantly contain highly imbalanced numbers of benign versus malignant samples. In turn, this imbalance introduces substantial bias in classification models due to the statistical dominance of the majority class. To address this issue, we introduce a deep clustering approach based on the latent-space embedding of dermoscopic images. Clustering is achieved using a novel center-oriented margin-free triplet loss (COM-Triplet) enforced on image embeddings from a convolutional neural network backbone. The proposed method aims to form maximally-separated cluster centers as opposed to minimizing classification error, so it is less sensitive to class imbalance. To avoid the need for labeled data, we further propose to implement COM-Triplet based on pseudo-labels generated by a Gaussian mixture model. Comprehensive experiments show that deep clustering with COM-Triplet loss outperforms clustering with triplet loss, and competing classifiers in both supervised and unsupervised settings.
Semantic Structure and Interpretability of Word Embeddings
May 16, 2018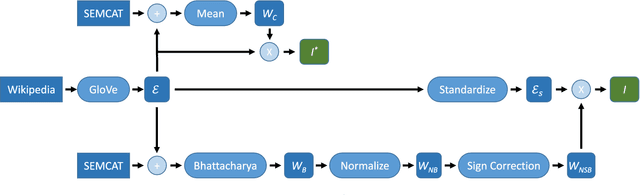
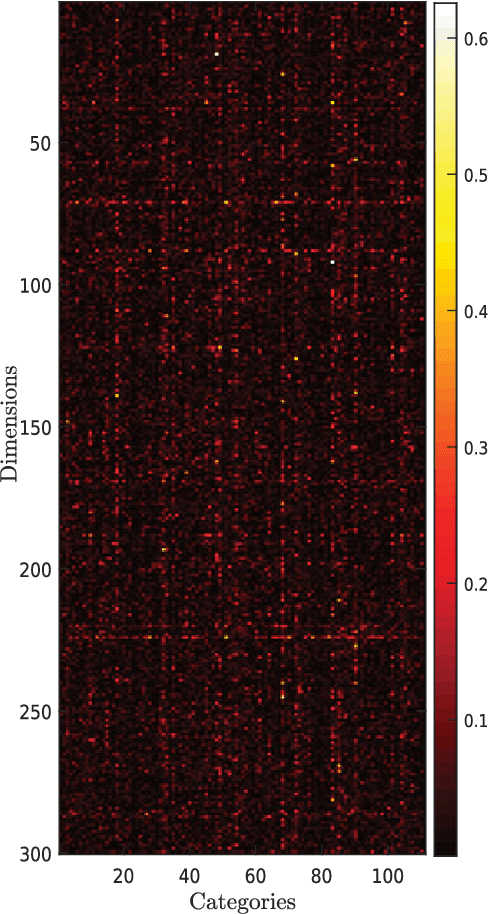
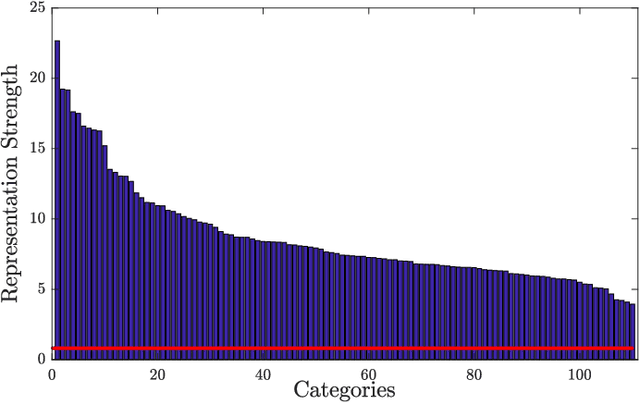
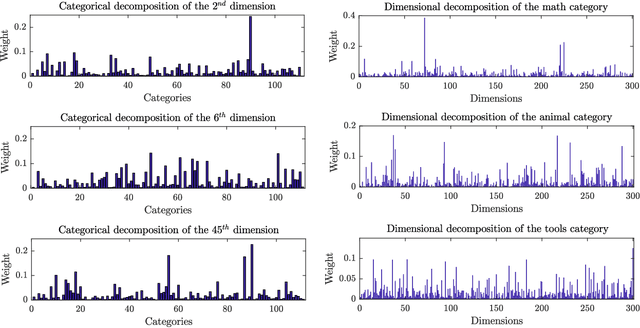
Abstract:Dense word embeddings, which encode semantic meanings of words to low dimensional vector spaces have become very popular in natural language processing (NLP) research due to their state-of-the-art performances in many NLP tasks. Word embeddings are substantially successful in capturing semantic relations among words, so a meaningful semantic structure must be present in the respective vector spaces. However, in many cases, this semantic structure is broadly and heterogeneously distributed across the embedding dimensions, which makes interpretation a big challenge. In this study, we propose a statistical method to uncover the latent semantic structure in the dense word embeddings. To perform our analysis we introduce a new dataset (SEMCAT) that contains more than 6500 words semantically grouped under 110 categories. We further propose a method to quantify the interpretability of the word embeddings; the proposed method is a practical alternative to the classical word intrusion test that requires human intervention.
* 11 Pages, 8 Figures, accepted by IEEE/ACM Transactions on Audio, Speech, and Language Processing
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge