Mehmet Orman
JointNET: A Deep Model for Predicting Active Sacroiliitis from Sacroiliac Joint Radiography
Jan 27, 2023Abstract:Purpose: To develop a deep learning model that predicts active inflammation from sacroiliac joint radiographs and to compare the success with radiologists. Materials and Methods: A total of 1,537 (augmented 1752) grade 0 SIJs of 768 patients were retrospectively analyzed. Gold-standard MRI exams showed active inflammation in 330 joints according to ASAS criteria. A convolutional neural network model (JointNET) was developed to detect MRI-based active inflammation labels solely based on radiographs. Two radiologists blindly evaluated the radiographs for comparison. Python, PyTorch, and SPSS were used for analyses. P<0.05 was considered statistically significant. Results: JointNET differentiated active inflammation from radiographs with a mean AUROC of 89.2 (95% CI:86.8%, 91.7%). The sensitivity was 69.0% (95% CI:65.3%, 72.7%) and specificity 90.4% (95% CI:87.8 % 92.9%). The mean accuracy was 90.2% (95% CI: 87.6%, 92.8%). The positive predictive value was 74.6% (95% CI: 72.5%, 76.7%) and negative predictive value was 87.9% (95% CI: 85.4%, 90.5%) when prevalence was considered 1%. Statistical analyses showed a significant difference between active inflammation and healthy groups (p<0.05). Radiologists accuracies were less than 65% to discriminate active inflammation from sacroiliac joint radiographs. Conclusion: JointNET successfully predicts active inflammation from sacroiliac joint radiographs, with superior performance to human observers.
Machine Learning Based Radiomics for Glial Tumor Classification and Comparison with Volumetric Analysis
Aug 13, 2022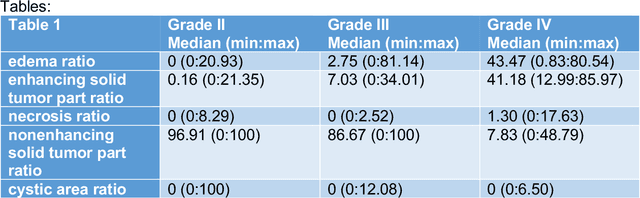
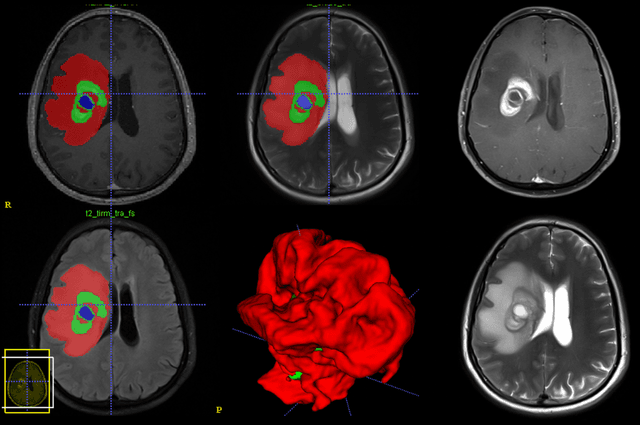
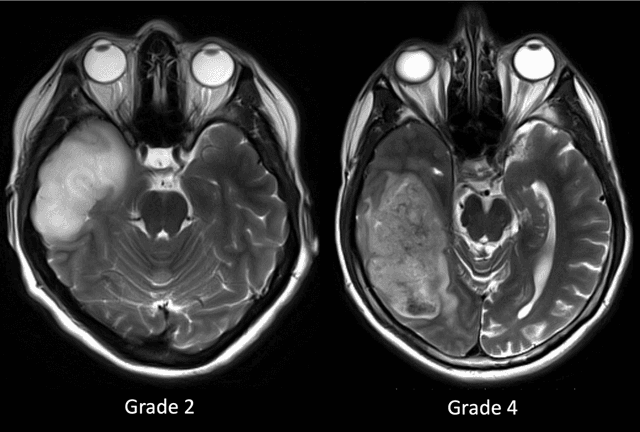
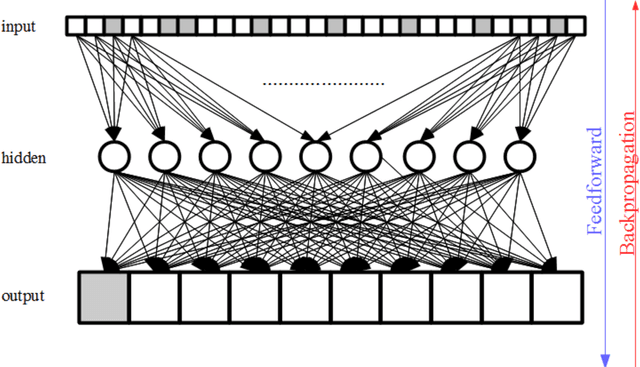
Abstract:Purpose; The purpose of this study is to classify glial tumors into grade II, III and IV categories noninvasively by application of machine learning to multi-modal MRI features in comparison with volumetric analysis. Methods; We retrospectively studied 57 glioma patients with pre and postcontrast T1 weighted, T2 weighted, FLAIR images, and ADC maps acquired on a 3T MRI. The tumors were segmented into enhancing and nonenhancing portions, tumor necrosis, cyst and edema using semiautomated segmentation of ITK-SNAP open source tool. We measured total tumor volume, enhancing-nonenhancing tumor, edema, necrosis volume and the ratios to the total tumor volume. Training of a support vector machine (SVM) classifier and artificial neural network (ANN) was performed with labeled data designed to answer the question of interest. Specificity, sensitivity, and AUC of the predictions were computed by means of ROC analysis. Differences in continuous measures between groups were assessed by using Kruskall Wallis, with post hoc Dunn correction for multiple comparisons. Results; When we compared the volume ratios between groups, there was statistically significant difference between grade IV and grade II-III glial tumors. Edema and tumor necrosis volume ratios for grade IV glial tumors were higher than that of grade II and III. Volumetric ratio analysis could not distinguish grade II and III tumors successfully. However, SVM and ANN correctly classified each group with accuracies up to 98% and 96%. Conclusion; Application of machine learning methods to MRI features can be used to classify brain tumors noninvasively and more readily in clinical settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge