Ashok Srinivasan
Machine Learning Based Radiomics for Glial Tumor Classification and Comparison with Volumetric Analysis
Aug 13, 2022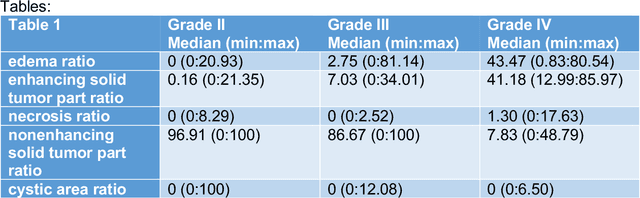
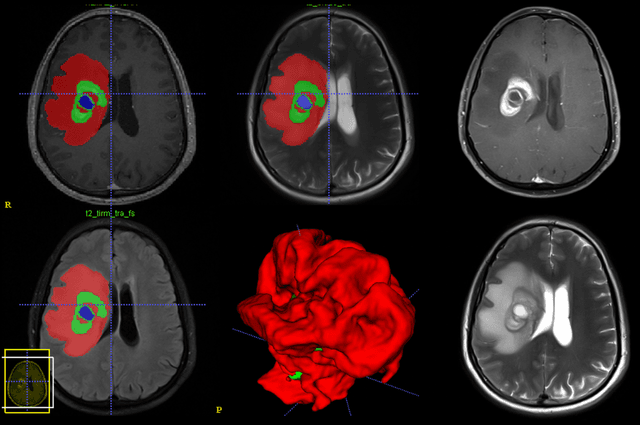
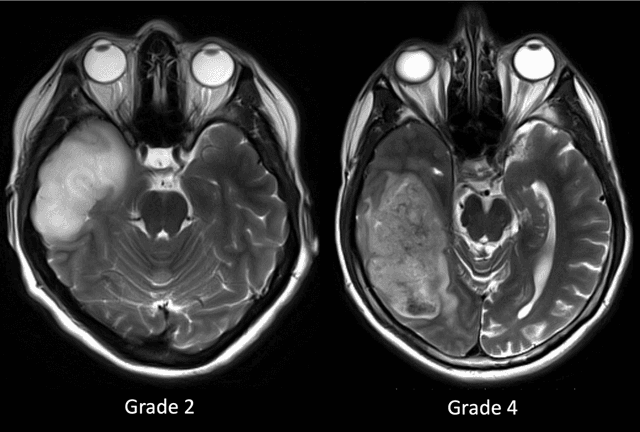
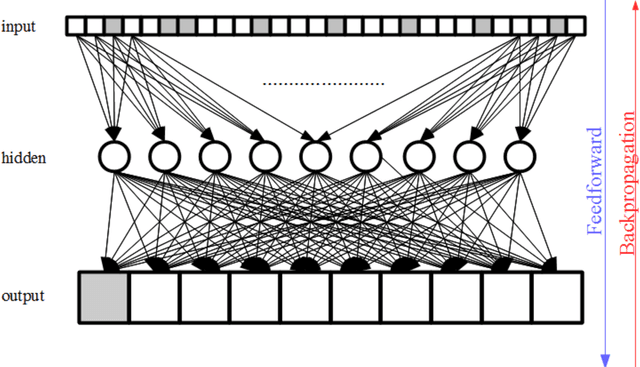
Abstract:Purpose; The purpose of this study is to classify glial tumors into grade II, III and IV categories noninvasively by application of machine learning to multi-modal MRI features in comparison with volumetric analysis. Methods; We retrospectively studied 57 glioma patients with pre and postcontrast T1 weighted, T2 weighted, FLAIR images, and ADC maps acquired on a 3T MRI. The tumors were segmented into enhancing and nonenhancing portions, tumor necrosis, cyst and edema using semiautomated segmentation of ITK-SNAP open source tool. We measured total tumor volume, enhancing-nonenhancing tumor, edema, necrosis volume and the ratios to the total tumor volume. Training of a support vector machine (SVM) classifier and artificial neural network (ANN) was performed with labeled data designed to answer the question of interest. Specificity, sensitivity, and AUC of the predictions were computed by means of ROC analysis. Differences in continuous measures between groups were assessed by using Kruskall Wallis, with post hoc Dunn correction for multiple comparisons. Results; When we compared the volume ratios between groups, there was statistically significant difference between grade IV and grade II-III glial tumors. Edema and tumor necrosis volume ratios for grade IV glial tumors were higher than that of grade II and III. Volumetric ratio analysis could not distinguish grade II and III tumors successfully. However, SVM and ANN correctly classified each group with accuracies up to 98% and 96%. Conclusion; Application of machine learning methods to MRI features can be used to classify brain tumors noninvasively and more readily in clinical settings.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
High-resolution home location prediction from tweets using deep learning with dynamic structure
Feb 03, 2019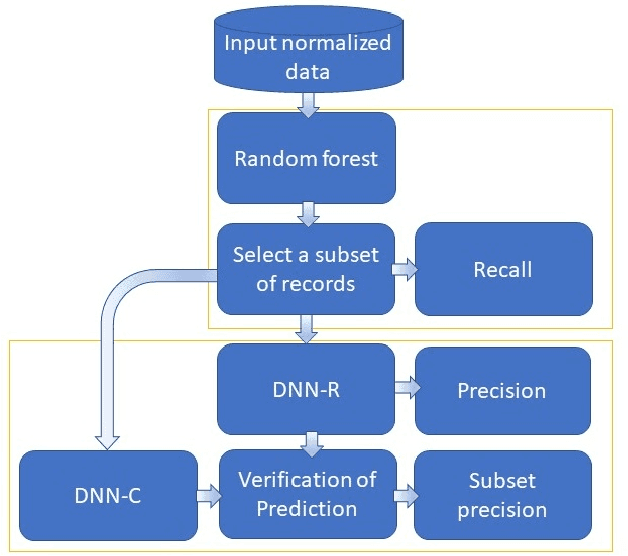

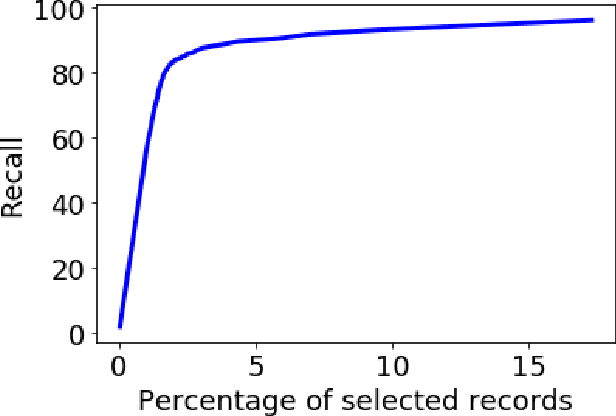
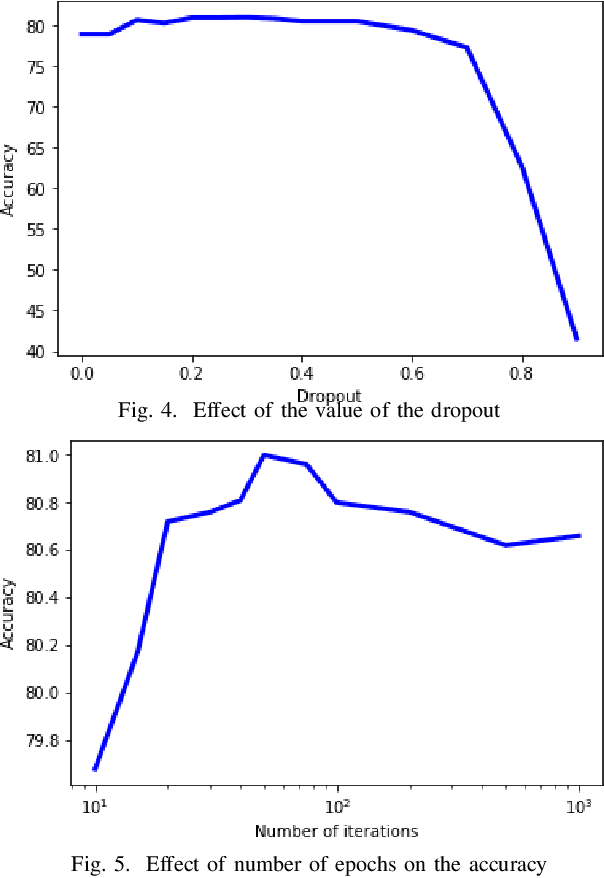
Abstract:High-resolution prediction of the home location of people has applications in diverse fields, including agriculture, transportation, and public health. The goal here is to obtain an accurate estimate of home locations of a sufficiently large subset of the population to subsequently use in models for the application domain. Conventional data sources, such as census and surveys, have a substantial time-lag and cannot capture seasonal trends. There has been much recent interest in the use of social media data to overcome this limitation. However, this data is usually sparse, noisy and user's home location is just one of several check-in locations. Due to these constraints, much of previous research has aimed at a coarse spatial resolution, such as at the time zone, state, and city levels. This is inadequate for important applications. For example, vector control to prevent epidemics would benefit from 200m resolution. Recent work has used a Support Vector Classifier on Twitter meta-data for such resolution, obtaining 70% accuracy for a 76% subset of the test population with 100m resolution. In contrast, we developed a deep learning model for this problem, applying a dynamic structure consisting of a random forest in the first phase and two fully connected deep neural networks in the second phase. We obtained over 90% on 30% subset of the test population. Given the large user base for Twitter, this is a sufficiently large subset for use in the modeling applications that we target. We believe that ours is the highest accuracy obtained for high-resolution home location prediction from Twitter data for both the entire sample and for its subsets. The primary contribution of this work lies in developing a deep-learning solution that uses a dynamic structure to deal with sparse and noisy social media data to yield accurate high resolution home locations from Twitter data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge