Daniel B. Ennis
Towards Universal Learning-based Model for Cardiac Image Reconstruction: Summary of the CMRxRecon2024 Challenge
Mar 05, 2025Abstract:Cardiovascular magnetic resonance (CMR) offers diverse imaging contrasts for assessment of cardiac function and tissue characterization. However, acquiring each single CMR modality is often time-consuming, and comprehensive clinical protocols require multiple modalities with various sampling patterns, further extending the overall acquisition time and increasing susceptibility to motion artifacts. Existing deep learning-based reconstruction methods are often designed for specific acquisition parameters, which limits their ability to generalize across a variety of scan scenarios. As part of the CMRxRecon Series, the CMRxRecon2024 challenge provides diverse datasets encompassing multi-modality multi-view imaging with various sampling patterns, and a platform for the international community to develop and benchmark reconstruction solutions in two well-crafted tasks. Task 1 is a modality-universal setting, evaluating the out-of-distribution generalization of the reconstructed model, while Task 2 follows sampling-universal setting assessing the one-for-all adaptability of the universal model. Main contributions include providing the first and largest publicly available multi-modality, multi-view cardiac k-space dataset; developing a benchmarking platform that simulates clinical acceleration protocols, with a shared code library and tutorial for various k-t undersampling patterns and data processing; giving technical insights of enhanced data consistency based on physic-informed networks and adaptive prompt-learning embedding to be versatile to different clinical settings; additional finding on evaluation metrics to address the limitations of conventional ground-truth references in universal reconstruction tasks.
On the Foundation Model for Cardiac MRI Reconstruction
Nov 15, 2024



Abstract:In recent years, machine learning (ML) based reconstruction has been widely investigated and employed in cardiac magnetic resonance (CMR) imaging. ML-based reconstructions can deliver clinically acceptable image quality under substantially accelerated scans. ML-based reconstruction, however, also requires substantial data and computational time to train the neural network, which is often optimized for a fixed acceleration rate or image contrast. In practice, imaging parameters are often tuned to best suit the diagnosis, which may differ from the training data. This can result in degraded image quality, and multiple trained networks are needed to fulfill the clinical demands. In this study, we propose a foundation model that uses adaptive unrolling, channel-shifting, and Pattern and Contrast-Prompt-UNet (PCP-UNet) to tackle the problem. In particular, the undersampled data goes through a different number of unrolled iterations according to its acceleration rate. Channel-shifting improves reconstructed data quality. The PCP-UNet is equipped with an image contrast and sampling pattern prompt. In vivo CMR experiments were performed using mixed combinations of image contrasts, acceleration rates, and (under)sampling patterns. The proposed foundation model has significantly improved image quality for a wide range of CMR protocols and outperforms the conventional ML-based method.
SDF4CHD: Generative Modeling of Cardiac Anatomies with Congenital Heart Defects
Nov 08, 2023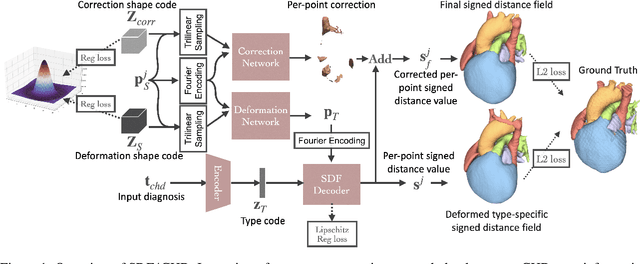
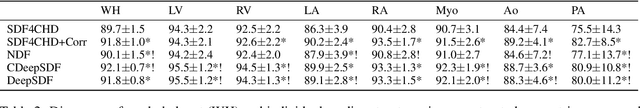
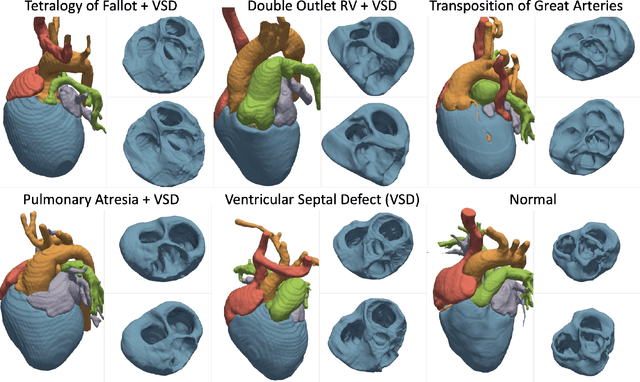
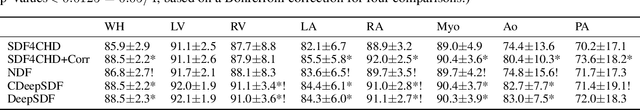
Abstract:Congenital heart disease (CHD) encompasses a spectrum of cardiovascular structural abnormalities, often requiring customized treatment plans for individual patients. Computational modeling and analysis of these unique cardiac anatomies can improve diagnosis and treatment planning and may ultimately lead to improved outcomes. Deep learning (DL) methods have demonstrated the potential to enable efficient treatment planning by automating cardiac segmentation and mesh construction for patients with normal cardiac anatomies. However, CHDs are often rare, making it challenging to acquire sufficiently large patient cohorts for training such DL models. Generative modeling of cardiac anatomies has the potential to fill this gap via the generation of virtual cohorts; however, prior approaches were largely designed for normal anatomies and cannot readily capture the significant topological variations seen in CHD patients. Therefore, we propose a type- and shape-disentangled generative approach suitable to capture the wide spectrum of cardiac anatomies observed in different CHD types and synthesize differently shaped cardiac anatomies that preserve the unique topology for specific CHD types. Our DL approach represents generic whole heart anatomies with CHD type-specific abnormalities implicitly using signed distance fields (SDF) based on CHD type diagnosis, which conveniently captures divergent anatomical variations across different types and represents meaningful intermediate CHD states. To capture the shape-specific variations, we then learn invertible deformations to morph the learned CHD type-specific anatomies and reconstruct patient-specific shapes. Our approach has the potential to augment the image-segmentation pairs for rarer CHD types for cardiac segmentation and generate cohorts of CHD cardiac meshes for computational simulation.
Coil Sketching for computationally-efficient MR iterative reconstruction
May 10, 2023Abstract:Purpose: Parallel imaging and compressed sensing reconstructions of large MRI datasets often have a prohibitive computational cost that bottlenecks clinical deployment, especially for 3D non-Cartesian acquisitions. One common approach is to reduce the number of coil channels actively used during reconstruction as in coil compression. While effective for Cartesian imaging, coil compression inherently loses signal energy, producing shading artifacts that compromise image quality for 3D non-Cartesian imaging. We propose coil sketching, a general and versatile method for computationally-efficient iterative MR image reconstruction. Theory and Methods: We based our method on randomized sketching algorithms, a type of large-scale optimization algorithms well established in the fields of machine learning and big data analysis. We adapt the sketching theory to the MRI reconstruction problem via a structured sketching matrix that, similar to coil compression, reduces the number of coils concurrently used during reconstruction, but unlike coil compression, is able to leverage energy from all coils. Results: First, we performed ablation experiments to validate the sketching matrix design on both Cartesian and non-Cartesian datasets. The resulting design yielded both improved computational efficiency and preserved signal-to-noise ratio (SNR) as measured by the inverse g-factor. Then, we verified the efficacy of our approach on high-dimensional non-Cartesian 3D cones datasets, where coil sketching yielded up to three-fold faster reconstructions with equivalent image quality. Conclusion: Coil sketching is a general and versatile reconstruction framework for computationally fast and memory-efficient reconstruction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge