Christopher M. Sandino
Coil Sketching for computationally-efficient MR iterative reconstruction
May 10, 2023Abstract:Purpose: Parallel imaging and compressed sensing reconstructions of large MRI datasets often have a prohibitive computational cost that bottlenecks clinical deployment, especially for 3D non-Cartesian acquisitions. One common approach is to reduce the number of coil channels actively used during reconstruction as in coil compression. While effective for Cartesian imaging, coil compression inherently loses signal energy, producing shading artifacts that compromise image quality for 3D non-Cartesian imaging. We propose coil sketching, a general and versatile method for computationally-efficient iterative MR image reconstruction. Theory and Methods: We based our method on randomized sketching algorithms, a type of large-scale optimization algorithms well established in the fields of machine learning and big data analysis. We adapt the sketching theory to the MRI reconstruction problem via a structured sketching matrix that, similar to coil compression, reduces the number of coils concurrently used during reconstruction, but unlike coil compression, is able to leverage energy from all coils. Results: First, we performed ablation experiments to validate the sketching matrix design on both Cartesian and non-Cartesian datasets. The resulting design yielded both improved computational efficiency and preserved signal-to-noise ratio (SNR) as measured by the inverse g-factor. Then, we verified the efficacy of our approach on high-dimensional non-Cartesian 3D cones datasets, where coil sketching yielded up to three-fold faster reconstructions with equivalent image quality. Conclusion: Coil sketching is a general and versatile reconstruction framework for computationally fast and memory-efficient reconstruction.
MAEEG: Masked Auto-encoder for EEG Representation Learning
Oct 27, 2022



Abstract:Decoding information from bio-signals such as EEG, using machine learning has been a challenge due to the small data-sets and difficulty to obtain labels. We propose a reconstruction-based self-supervised learning model, the masked auto-encoder for EEG (MAEEG), for learning EEG representations by learning to reconstruct the masked EEG features using a transformer architecture. We found that MAEEG can learn representations that significantly improve sleep stage classification (~5% accuracy increase) when only a small number of labels are given. We also found that input sample lengths and different ways of masking during reconstruction-based SSL pretraining have a huge effect on downstream model performance. Specifically, learning to reconstruct a larger proportion and more concentrated masked signal results in better performance on sleep classification. Our findings provide insight into how reconstruction-based SSL could help representation learning for EEG.
Memory-efficient Learning for High-Dimensional MRI Reconstruction
Mar 06, 2021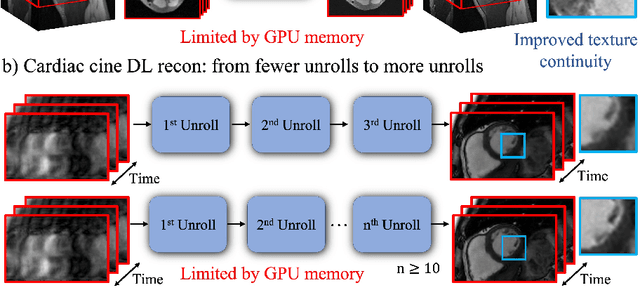



Abstract:Deep learning (DL) based unrolled reconstructions have shown state-of-the-art performance for under-sampled magnetic resonance imaging (MRI). Similar to compressed sensing, DL can leverage high-dimensional data (e.g. 3D, 2D+time, 3D+time) to further improve performance. However, network size and depth are currently limited by the GPU memory required for backpropagation. Here we use a memory-efficient learning (MEL) framework which favorably trades off storage with a manageable increase in computation during training. Using MEL with multi-dimensional data, we demonstrate improved image reconstruction performance for in-vivo 3D MRI and 2D+time cardiac cine MRI. MEL uses far less GPU memory while marginally increasing the training time, which enables new applications of DL to high-dimensional MRI.
Accelerating cardiac cine MRI beyond compressed sensing using DL-ESPIRiT
Nov 13, 2019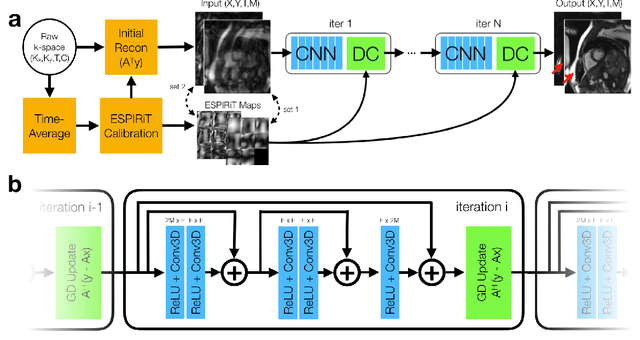
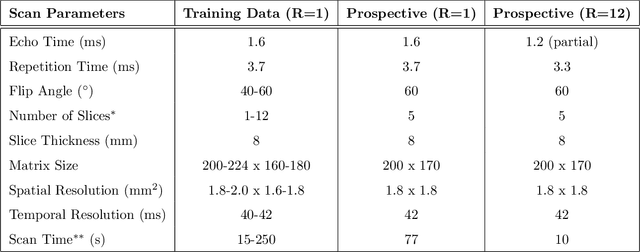
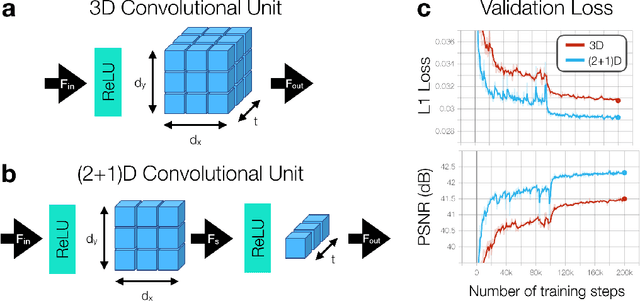
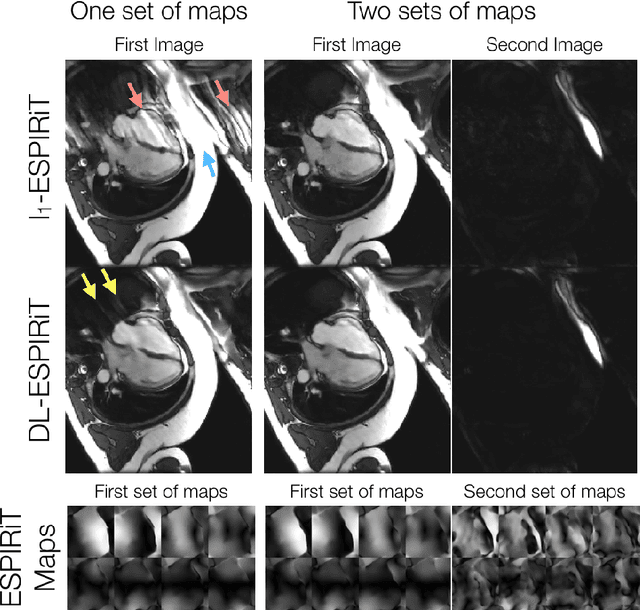
Abstract:A novel neural network architecture, known as DL-ESPIRiT, is proposed to reconstruct rapidly acquired cardiac MRI data without field-of-view limitations which are present in previously proposed deep learning-based reconstruction frameworks. Additionally, a novel convolutional neural network based on separable 3D convolutions is integrated into DL-ESPIRiT to more efficiently learn spatiotemporal priors for dynamic image reconstruction. The network is trained on fully-sampled 2D cardiac cine datasets collected from eleven healthy volunteers with IRB approval. DL-ESPIRiT is compared against a state-of-the-art parallel imaging and compressed sensing method known as $l_1$-ESPIRiT. The reconstruction accuracy of both methods is evaluated on retrospectively undersampled datasets (R=12) with respect to standard image quality metrics as well as automatic deep learning-based segmentations of left ventricular volumes. Feasibility of this approach is demonstrated in reconstructions of prospectively undersampled data which were acquired in a single heartbeat per slice.
Reconstruction of Undersampled 3D Non-Cartesian Image-Based Navigators for Coronary MRA Using an Unrolled Deep Learning Model
Oct 24, 2019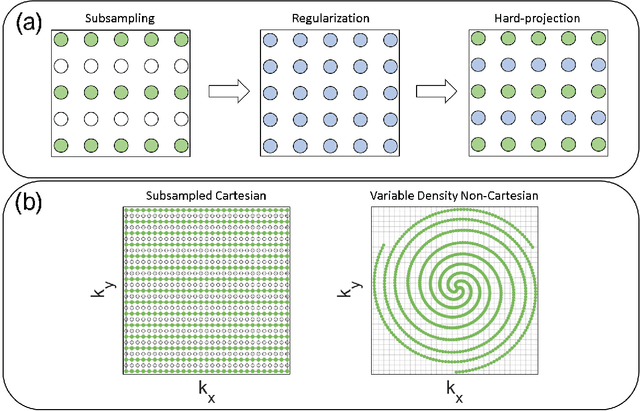

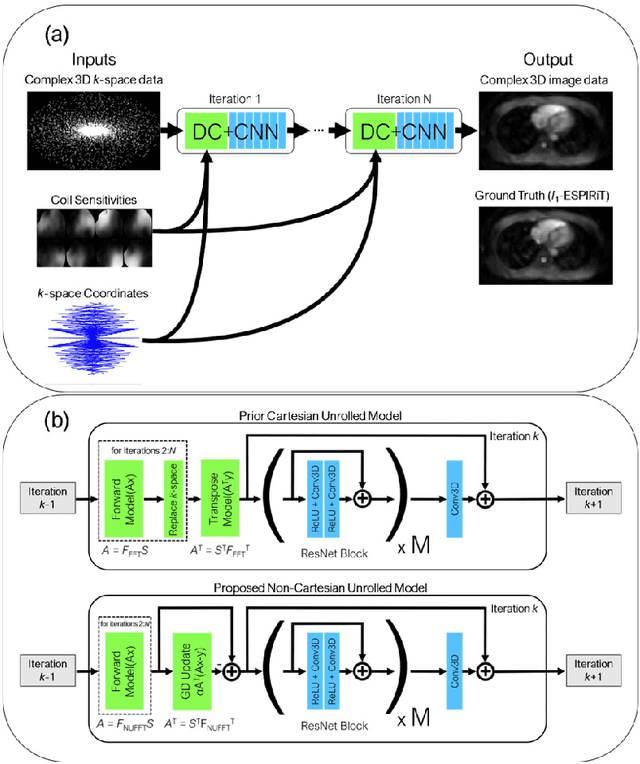
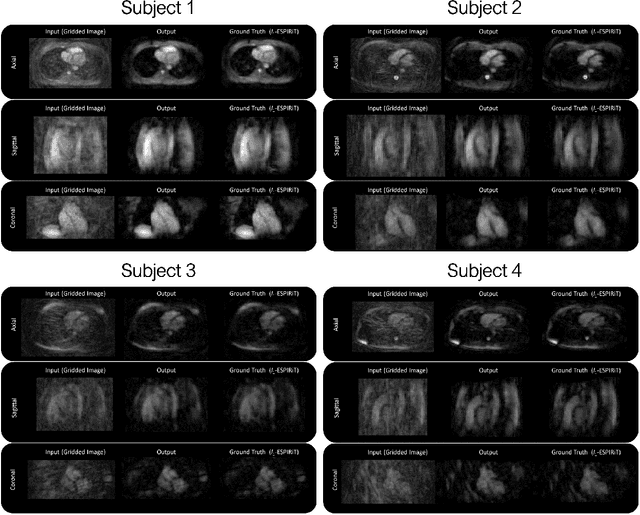
Abstract:Purpose: To rapidly reconstruct undersampled 3D non-Cartesian image-based navigators (iNAVs) using an unrolled deep learning (DL) model for non-rigid motion correction in coronary magnetic resonance angiography (CMRA). Methods: An unrolled network is trained to reconstruct beat-to-beat 3D iNAVs acquired as part of a CMRA sequence. The unrolled model incorporates a non-uniform FFT operator to perform the data consistency operation, and the regularization term is learned by a convolutional neural network (CNN) based on the proximal gradient descent algorithm. The training set includes 6,000 3D iNAVs acquired from 7 different subjects and 11 scans using a variable-density (VD) cones trajectory. For testing, 3D iNAVs from 4 additional subjects are reconstructed using the unrolled model. To validate reconstruction accuracy, global and localized motion estimates from DL model-based 3D iNAVs are compared with those extracted from 3D iNAVs reconstructed with $\textit{l}_{1}$-ESPIRiT. Then, the high-resolution coronary MRA images motion corrected with autofocusing using the $\textit{l}_{1}$-ESPIRiT and DL model-based 3D iNAVs are assessed for differences. Results: 3D iNAVs reconstructed using the DL model-based approach and conventional $\textit{l}_{1}$-ESPIRiT generate similar global and localized motion estimates and provide equivalent coronary image quality. Reconstruction with the unrolled network completes in a fraction of the time compared to CPU and GPU implementations of $\textit{l}_{1}$-ESPIRiT (20x and 3x speed increases, respectively). Conclusion: We have developed a deep neural network architecture to reconstruct undersampled 3D non-Cartesian VD cones iNAVs. Our approach decreases reconstruction time for 3D iNAVs, while preserving the accuracy of non-rigid motion information offered by them for correction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge