Jonathan I. Tamir
Non-rigid Motion Correction for MRI Reconstruction via Coarse-To-Fine Diffusion Models
May 21, 2025



Abstract:Magnetic Resonance Imaging (MRI) is highly susceptible to motion artifacts due to the extended acquisition times required for k-space sampling. These artifacts can compromise diagnostic utility, particularly for dynamic imaging. We propose a novel alternating minimization framework that leverages a bespoke diffusion model to jointly reconstruct and correct non-rigid motion-corrupted k-space data. The diffusion model uses a coarse-to-fine denoising strategy to capture large overall motion and reconstruct the lower frequencies of the image first, providing a better inductive bias for motion estimation than that of standard diffusion models. We demonstrate the performance of our approach on both real-world cine cardiac MRI datasets and complex simulated rigid and non-rigid deformations, even when each motion state is undersampled by a factor of 64x. Additionally, our method is agnostic to sampling patterns, anatomical variations, and MRI scanning protocols, as long as some low frequency components are sampled during each motion state.
Diffusion Probabilistic Generative Models for Accelerated, in-NICU Permanent Magnet Neonatal MRI
May 21, 2025Abstract:Purpose: Magnetic Resonance Imaging (MRI) enables non-invasive assessment of brain abnormalities during early life development. Permanent magnet scanners operating in the neonatal intensive care unit (NICU) facilitate MRI of sick infants, but have long scan times due to lower signal-to-noise ratios (SNR) and limited receive coils. This work accelerates in-NICU MRI with diffusion probabilistic generative models by developing a training pipeline accounting for these challenges. Methods: We establish a novel training dataset of clinical, 1 Tesla neonatal MR images in collaboration with Aspect Imaging and Sha'are Zedek Medical Center. We propose a pipeline to handle the low quantity and SNR of our real-world dataset (1) modifying existing network architectures to support varying resolutions; (2) training a single model on all data with learned class embedding vectors; (3) applying self-supervised denoising before training; and (4) reconstructing by averaging posterior samples. Retrospective under-sampling experiments, accounting for signal decay, evaluated each item of our proposed methodology. A clinical reader study with practicing pediatric neuroradiologists evaluated our proposed images reconstructed from 1.5x under-sampled data. Results: Combining all data, denoising pre-training, and averaging posterior samples yields quantitative improvements in reconstruction. The generative model decouples the learned prior from the measurement model and functions at two acceleration rates without re-training. The reader study suggests that proposed images reconstructed from approximately 1.5x under-sampled data are adequate for clinical use. Conclusion: Diffusion probabilistic generative models applied with the proposed pipeline to handle challenging real-world datasets could reduce scan time of in-NICU neonatal MRI.
Double Blind Imaging with Generative Modeling
Mar 27, 2025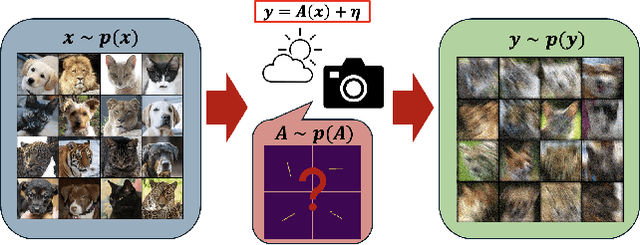
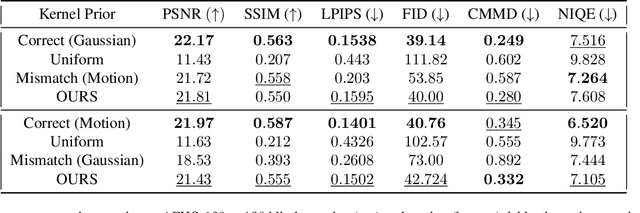
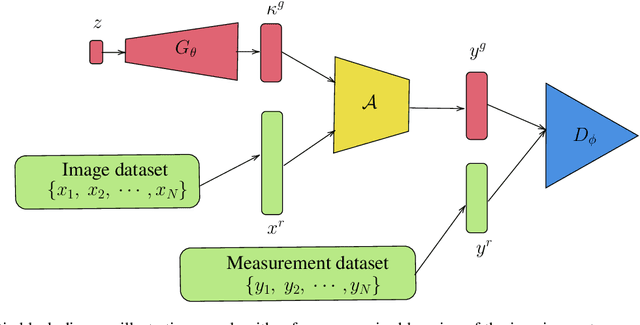
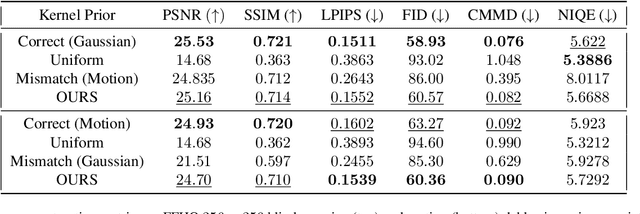
Abstract:Blind inverse problems in imaging arise from uncertainties in the system used to collect (noisy) measurements of images. Recovering clean images from these measurements typically requires identifying the imaging system, either implicitly or explicitly. A common solution leverages generative models as priors for both the images and the imaging system parameters (e.g., a class of point spread functions). To learn these priors in a straightforward manner requires access to a dataset of clean images as well as samples of the imaging system. We propose an AmbientGAN-based generative technique to identify the distribution of parameters in unknown imaging systems, using only unpaired clean images and corrupted measurements. This learned distribution can then be used in model-based recovery algorithms to solve blind inverse problems such as blind deconvolution. We successfully demonstrate our technique for learning Gaussian blur and motion blur priors from noisy measurements and show their utility in solving blind deconvolution with diffusion posterior sampling.
Enhancing Deep Learning-Driven Multi-Coil MRI Reconstruction via Self-Supervised Denoising
Nov 19, 2024Abstract:We examine the effect of incorporating self-supervised denoising as a pre-processing step for training deep learning (DL) based reconstruction methods on data corrupted by Gaussian noise. K-space data employed for training are typically multi-coil and inherently noisy. Although DL-based reconstruction methods trained on fully sampled data can enable high reconstruction quality, obtaining large, noise-free datasets is impractical. We leverage Generalized Stein's Unbiased Risk Estimate (GSURE) for denoising. We evaluate two DL-based reconstruction methods: Diffusion Probabilistic Models (DPMs) and Model-Based Deep Learning (MoDL). We evaluate the impact of denoising on the performance of these DL-based methods in solving accelerated multi-coil magnetic resonance imaging (MRI) reconstruction. The experiments were carried out on T2-weighted brain and fat-suppressed proton-density knee scans. We observed that self-supervised denoising enhances the quality and efficiency of MRI reconstructions across various scenarios. Specifically, employing denoised images rather than noisy counterparts when training DL networks results in lower normalized root mean squared error (NRMSE), higher structural similarity index measure (SSIM) and peak signal-to-noise ratio (PSNR) across different SNR levels, including 32dB, 22dB, and 12dB for T2-weighted brain data, and 24dB, 14dB, and 4dB for fat-suppressed knee data. Overall, we showed that denoising is an essential pre-processing technique capable of improving the efficacy of DL-based MRI reconstruction methods under diverse conditions. By refining the quality of input data, denoising can enable the training of more effective DL networks, potentially bypassing the need for noise-free reference MRI scans.
Accelerated, Robust Lower-Field Neonatal MRI with Generative Models
Oct 28, 2024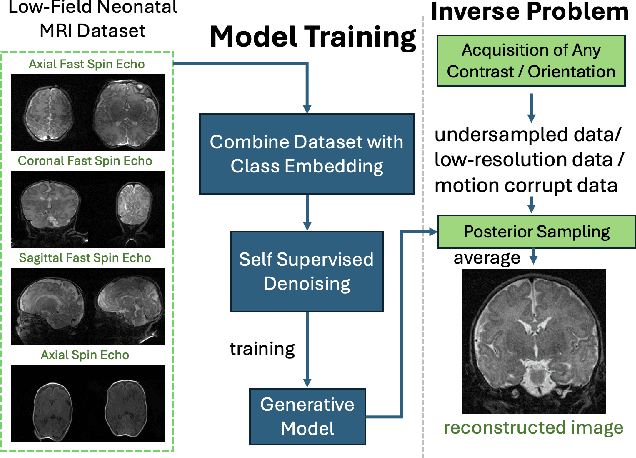
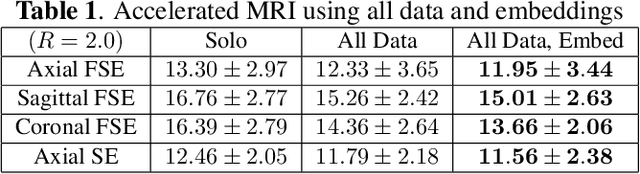
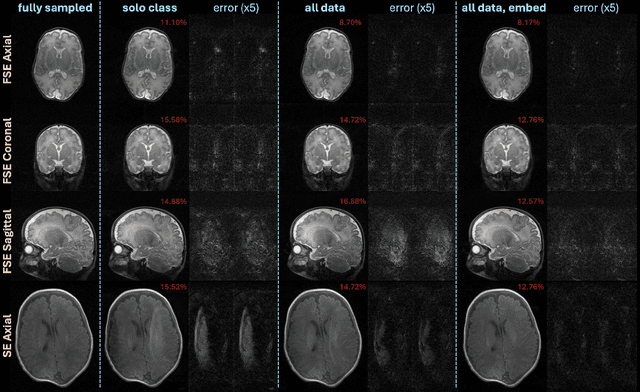
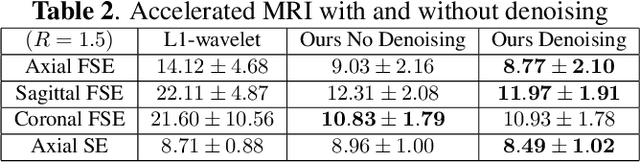
Abstract:Neonatal Magnetic Resonance Imaging (MRI) enables non-invasive assessment of potential brain abnormalities during the critical phase of early life development. Recently, interest has developed in lower field (i.e., below 1.5 Tesla) MRI systems that trade-off magnetic field strength for portability and access in the neonatal intensive care unit (NICU). Unfortunately, lower-field neonatal MRI still suffers from long scan times and motion artifacts that can limit its clinical utility for neonates. This work improves motion robustness and accelerates lower field neonatal MRI through diffusion-based generative modeling and signal processing based motion modeling. We first gather a training dataset of clinical neonatal MRI images. Then we train a diffusion-based generative model to learn the statistical distribution of fully-sampled images by applying several signal processing methods to handle the lower signal-to-noise ratio and lower quality of our MRI images. Finally, we present experiments demonstrating the utility of our generative model to improve reconstruction performance across two tasks: accelerated MRI and motion correction.
Evaluating Neural Networks for Early Maritime Threat Detection
Oct 26, 2024Abstract:We consider the task of classifying trajectories of boat activities as a proxy for assessing maritime threats. Previous approaches have considered entropy-based metrics for clustering boat activity into three broad categories: random walk, following, and chasing. Here, we comprehensively assess the accuracy of neural network-based approaches as alternatives to entropy-based clustering. We train four neural network models and compare them to shallow learning using synthetic data. We also investigate the accuracy of models as time steps increase and with and without rotated data. To improve test-time robustness, we normalize trajectories and perform rotation-based data augmentation. Our results show that deep networks can achieve a test-set accuracy of up to 100% on a full trajectory, with graceful degradation as the number of time steps decreases, outperforming entropy-based clustering.
Ambient Diffusion Posterior Sampling: Solving Inverse Problems with Diffusion Models trained on Corrupted Data
Mar 13, 2024



Abstract:We provide a framework for solving inverse problems with diffusion models learned from linearly corrupted data. Our method, Ambient Diffusion Posterior Sampling (A-DPS), leverages a generative model pre-trained on one type of corruption (e.g. image inpainting) to perform posterior sampling conditioned on measurements from a potentially different forward process (e.g. image blurring). We test the efficacy of our approach on standard natural image datasets (CelebA, FFHQ, and AFHQ) and we show that A-DPS can sometimes outperform models trained on clean data for several image restoration tasks in both speed and performance. We further extend the Ambient Diffusion framework to train MRI models with access only to Fourier subsampled multi-coil MRI measurements at various acceleration factors (R=2, 4, 6, 8). We again observe that models trained on highly subsampled data are better priors for solving inverse problems in the high acceleration regime than models trained on fully sampled data. We open-source our code and the trained Ambient Diffusion MRI models: https://github.com/utcsilab/ambient-diffusion-mri .
Optimizing Sampling Patterns for Compressed Sensing MRI with Diffusion Generative Models
Jun 05, 2023



Abstract:Diffusion-based generative models have been used as powerful priors for magnetic resonance imaging (MRI) reconstruction. We present a learning method to optimize sub-sampling patterns for compressed sensing multi-coil MRI that leverages pre-trained diffusion generative models. Crucially, during training we use a single-step reconstruction based on the posterior mean estimate given by the diffusion model and the MRI measurement process. Experiments across varying anatomies, acceleration factors, and pattern types show that sampling operators learned with our method lead to competitive, and in the case of 2D patterns, improved reconstructions compared to baseline patterns. Our method requires as few as five training images to learn effective sampling patterns.
Solving Inverse Problems with Score-Based Generative Priors learned from Noisy Data
May 02, 2023



Abstract:We present SURE-Score: an approach for learning score-based generative models using training samples corrupted by additive Gaussian noise. When a large training set of clean samples is available, solving inverse problems via score-based (diffusion) generative models trained on the underlying fully-sampled data distribution has recently been shown to outperform end-to-end supervised deep learning. In practice, such a large collection of training data may be prohibitively expensive to acquire in the first place. In this work, we present an approach for approximately learning a score-based generative model of the clean distribution, from noisy training data. We formulate and justify a novel loss function that leverages Stein's unbiased risk estimate to jointly denoise the data and learn the score function via denoising score matching, while using only the noisy samples. We demonstrate the generality of SURE-Score by learning priors and applying posterior sampling to ill-posed inverse problems in two practical applications from different domains: compressive wireless multiple-input multiple-output channel estimation and accelerated 2D multi-coil magnetic resonance imaging reconstruction, where we demonstrate competitive reconstruction performance when learning at signal-to-noise ratio values of 0 and 10 dB, respectively.
Conditional Score-Based Reconstructions for Multi-contrast MRI
Mar 26, 2023



Abstract:Magnetic resonance imaging (MRI) exam protocols consist of multiple contrast-weighted images of the same anatomy to emphasize different tissue properties. Due to the long acquisition times required to collect fully sampled k-space measurements, it is common to only collect a fraction of k-space for some, or all, of the scans and subsequently solve an inverse problem for each contrast to recover the desired image from sub-sampled measurements. Recently, there has been a push to further accelerate MRI exams using data-driven priors, and generative models in particular, to regularize the ill-posed inverse problem of image reconstruction. These methods have shown promising improvements over classical methods. However, many of the approaches neglect the multi-contrast nature of clinical MRI exams and treat each scan as an independent reconstruction. In this work we show that by learning a joint Bayesian prior over multi-contrast data with a score-based generative model we are able to leverage the underlying structure between multi-contrast images and thus improve image reconstruction fidelity over generative models that only reconstruct images of a single contrast.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge