Akshay Chaudhari
Department of Biomedical Data Science, Stanford University, California, USA, Department of Radiology, Stanford University, California, USA
Prompt Triage: Structured Optimization Enhances Vision-Language Model Performance on Medical Imaging Benchmarks
Nov 14, 2025Abstract:Vision-language foundation models (VLMs) show promise for diverse imaging tasks but often underperform on medical benchmarks. Prior efforts to improve performance include model finetuning, which requires large domain-specific datasets and significant compute, or manual prompt engineering, which is hard to generalize and often inaccessible to medical institutions seeking to deploy these tools. These challenges motivate interest in approaches that draw on a model's embedded knowledge while abstracting away dependence on human-designed prompts to enable scalable, weight-agnostic performance improvements. To explore this, we adapt the Declarative Self-improving Python (DSPy) framework for structured automated prompt optimization in medical vision-language systems through a comprehensive, formal evaluation. We implement prompting pipelines for five medical imaging tasks across radiology, gastroenterology, and dermatology, evaluating 10 open-source VLMs with four prompt optimization techniques. Optimized pipelines achieved a median relative improvement of 53% over zero-shot prompting baselines, with the largest gains ranging from 300% to 3,400% on tasks where zero-shot performance is low. These results highlight the substantial potential of applying automated prompt optimization to medical AI systems, demonstrating significant gains for vision-based applications requiring accurate clinical image interpretation. By reducing dependence on prompt design to elicit intended outputs, these techniques allow clinicians to focus on patient care and clinical decision-making. Furthermore, our experiments offer scalability and preserve data privacy, demonstrating performance improvement on open-source VLMs. We publicly release our evaluation pipelines to support reproducible research on specialized medical tasks, available at https://github.com/DaneshjouLab/prompt-triage-lab.
Agentic Systems in Radiology: Design, Applications, Evaluation, and Challenges
Oct 10, 2025Abstract:Building agents, systems that perceive and act upon their environment with a degree of autonomy, has long been a focus of AI research. This pursuit has recently become vastly more practical with the emergence of large language models (LLMs) capable of using natural language to integrate information, follow instructions, and perform forms of "reasoning" and planning across a wide range of tasks. With its multimodal data streams and orchestrated workflows spanning multiple systems, radiology is uniquely suited to benefit from agents that can adapt to context and automate repetitive yet complex tasks. In radiology, LLMs and their multimodal variants have already demonstrated promising performance for individual tasks such as information extraction and report summarization. However, using LLMs in isolation underutilizes their potential to support complex, multi-step workflows where decisions depend on evolving context from multiple information sources. Equipping LLMs with external tools and feedback mechanisms enables them to drive systems that exhibit a spectrum of autonomy, ranging from semi-automated workflows to more adaptive agents capable of managing complex processes. This review examines the design of such LLM-driven agentic systems, highlights key applications, discusses evaluation methods for planning and tool use, and outlines challenges such as error cascades, tool-use efficiency, and health IT integration.
MedHELM: Holistic Evaluation of Large Language Models for Medical Tasks
May 26, 2025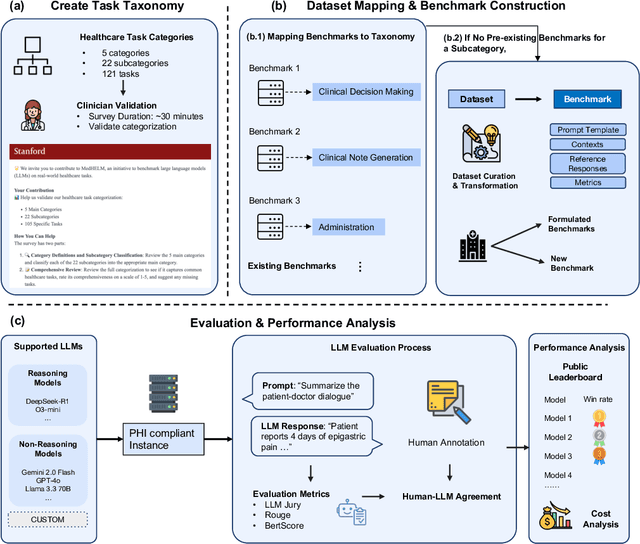
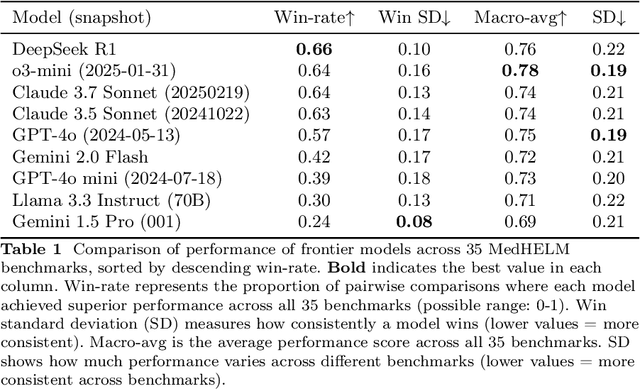
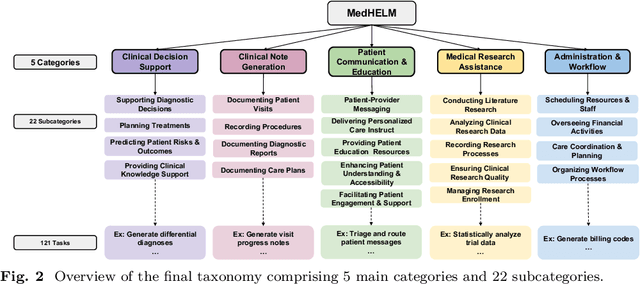
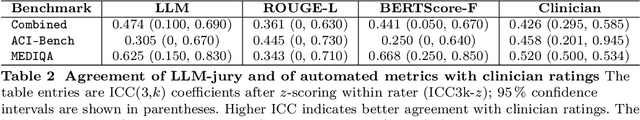
Abstract:While large language models (LLMs) achieve near-perfect scores on medical licensing exams, these evaluations inadequately reflect the complexity and diversity of real-world clinical practice. We introduce MedHELM, an extensible evaluation framework for assessing LLM performance for medical tasks with three key contributions. First, a clinician-validated taxonomy spanning 5 categories, 22 subcategories, and 121 tasks developed with 29 clinicians. Second, a comprehensive benchmark suite comprising 35 benchmarks (17 existing, 18 newly formulated) providing complete coverage of all categories and subcategories in the taxonomy. Third, a systematic comparison of LLMs with improved evaluation methods (using an LLM-jury) and a cost-performance analysis. Evaluation of 9 frontier LLMs, using the 35 benchmarks, revealed significant performance variation. Advanced reasoning models (DeepSeek R1: 66% win-rate; o3-mini: 64% win-rate) demonstrated superior performance, though Claude 3.5 Sonnet achieved comparable results at 40% lower estimated computational cost. On a normalized accuracy scale (0-1), most models performed strongly in Clinical Note Generation (0.73-0.85) and Patient Communication & Education (0.78-0.83), moderately in Medical Research Assistance (0.65-0.75), and generally lower in Clinical Decision Support (0.56-0.72) and Administration & Workflow (0.53-0.63). Our LLM-jury evaluation method achieved good agreement with clinician ratings (ICC = 0.47), surpassing both average clinician-clinician agreement (ICC = 0.43) and automated baselines including ROUGE-L (0.36) and BERTScore-F1 (0.44). Claude 3.5 Sonnet achieved comparable performance to top models at lower estimated cost. These findings highlight the importance of real-world, task-specific evaluation for medical use of LLMs and provides an open source framework to enable this.
Eyes Tell the Truth: GazeVal Highlights Shortcomings of Generative AI in Medical Imaging
Mar 26, 2025



Abstract:The demand for high-quality synthetic data for model training and augmentation has never been greater in medical imaging. However, current evaluations predominantly rely on computational metrics that fail to align with human expert recognition. This leads to synthetic images that may appear realistic numerically but lack clinical authenticity, posing significant challenges in ensuring the reliability and effectiveness of AI-driven medical tools. To address this gap, we introduce GazeVal, a practical framework that synergizes expert eye-tracking data with direct radiological evaluations to assess the quality of synthetic medical images. GazeVal leverages gaze patterns of radiologists as they provide a deeper understanding of how experts perceive and interact with synthetic data in different tasks (i.e., diagnostic or Turing tests). Experiments with sixteen radiologists revealed that 96.6% of the generated images (by the most recent state-of-the-art AI algorithm) were identified as fake, demonstrating the limitations of generative AI in producing clinically accurate images.
MedVAE: Efficient Automated Interpretation of Medical Images with Large-Scale Generalizable Autoencoders
Feb 20, 2025



Abstract:Medical images are acquired at high resolutions with large fields of view in order to capture fine-grained features necessary for clinical decision-making. Consequently, training deep learning models on medical images can incur large computational costs. In this work, we address the challenge of downsizing medical images in order to improve downstream computational efficiency while preserving clinically-relevant features. We introduce MedVAE, a family of six large-scale 2D and 3D autoencoders capable of encoding medical images as downsized latent representations and decoding latent representations back to high-resolution images. We train MedVAE autoencoders using a novel two-stage training approach with 1,052,730 medical images. Across diverse tasks obtained from 20 medical image datasets, we demonstrate that (1) utilizing MedVAE latent representations in place of high-resolution images when training downstream models can lead to efficiency benefits (up to 70x improvement in throughput) while simultaneously preserving clinically-relevant features and (2) MedVAE can decode latent representations back to high-resolution images with high fidelity. Our work demonstrates that large-scale, generalizable autoencoders can help address critical efficiency challenges in the medical domain. Our code is available at https://github.com/StanfordMIMI/MedVAE.
Explaining 3D Computed Tomography Classifiers with Counterfactuals
Feb 11, 2025


Abstract:Counterfactual explanations in medical imaging are critical for understanding the predictions made by deep learning models. We extend the Latent Shift counterfactual generation method from 2D applications to 3D computed tomography (CT) scans. We address the challenges associated with 3D data, such as limited training samples and high memory demands, by implementing a slice-based approach. This method leverages a 2D encoder trained on CT slices, which are subsequently combined to maintain 3D context. We demonstrate this technique on two models for clinical phenotype prediction and lung segmentation. Our approach is both memory-efficient and effective for generating interpretable counterfactuals in high-resolution 3D medical imaging.
Embedding-Driven Diversity Sampling to Improve Few-Shot Synthetic Data Generation
Jan 20, 2025



Abstract:Accurate classification of clinical text often requires fine-tuning pre-trained language models, a process that is costly and time-consuming due to the need for high-quality data and expert annotators. Synthetic data generation offers an alternative, though pre-trained models may not capture the syntactic diversity of clinical notes. We propose an embedding-driven approach that uses diversity sampling from a small set of real clinical notes to guide large language models in few-shot prompting, generating synthetic text that better reflects clinical syntax. We evaluated this method using the CheXpert dataset on a classification task, comparing it to random few-shot and zero-shot approaches. Using cosine similarity and a Turing test, our approach produced synthetic notes that more closely align with real clinical text. Our pipeline reduced the data needed to reach the 0.85 AUC cutoff by 40% for AUROC and 30% for AUPRC, while augmenting models with synthetic data improved AUROC by 57% and AUPRC by 68%. Additionally, our synthetic data was 0.9 times as effective as real data, a 60% improvement in value.
Best Practices for Large Language Models in Radiology
Dec 02, 2024



Abstract:At the heart of radiological practice is the challenge of integrating complex imaging data with clinical information to produce actionable insights. Nuanced application of language is key for various activities, including managing requests, describing and interpreting imaging findings in the context of clinical data, and concisely documenting and communicating the outcomes. The emergence of large language models (LLMs) offers an opportunity to improve the management and interpretation of the vast data in radiology. Despite being primarily general-purpose, these advanced computational models demonstrate impressive capabilities in specialized language-related tasks, even without specific training. Unlocking the potential of LLMs for radiology requires basic understanding of their foundations and a strategic approach to navigate their idiosyncrasies. This review, drawing from practical radiology and machine learning expertise and recent literature, provides readers insight into the potential of LLMs in radiology. It examines best practices that have so far stood the test of time in the rapidly evolving landscape of LLMs. This includes practical advice for optimizing LLM characteristics for radiology practices along with limitations, effective prompting, and fine-tuning strategies.
Foundation Models in Radiology: What, How, When, Why and Why Not
Nov 27, 2024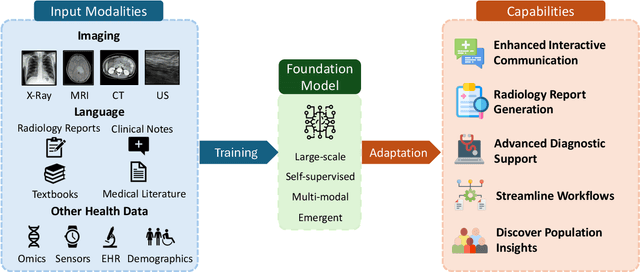
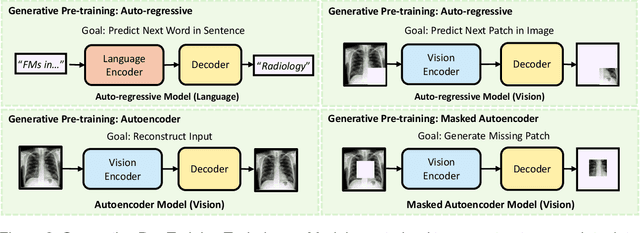
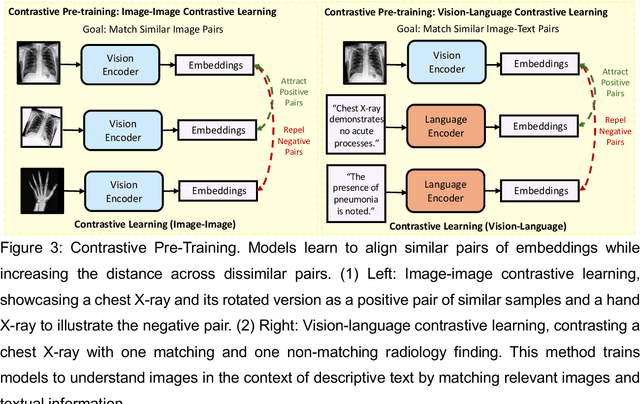
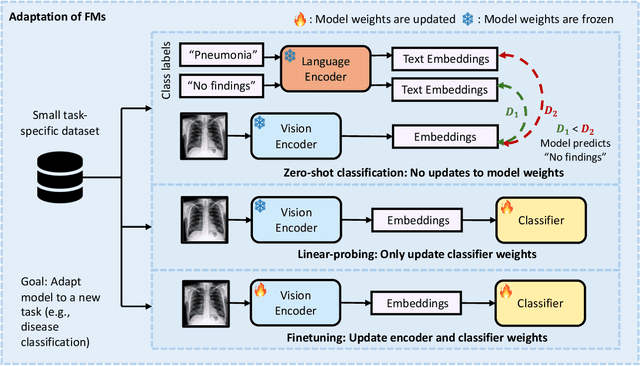
Abstract:Recent advances in artificial intelligence have witnessed the emergence of large-scale deep learning models capable of interpreting and generating both textual and imaging data. Such models, typically referred to as foundation models, are trained on extensive corpora of unlabeled data and demonstrate high performance across various tasks. Foundation models have recently received extensive attention from academic, industry, and regulatory bodies. Given the potentially transformative impact that foundation models can have on the field of radiology, this review aims to establish a standardized terminology concerning foundation models, with a specific focus on the requirements of training data, model training paradigms, model capabilities, and evaluation strategies. We further outline potential pathways to facilitate the training of radiology-specific foundation models, with a critical emphasis on elucidating both the benefits and challenges associated with such models. Overall, we envision that this review can unify technical advances and clinical needs in the training of foundation models for radiology in a safe and responsible manner, for ultimately benefiting patients, providers, and radiologists.
Evaluating and Improving the Effectiveness of Synthetic Chest X-Rays for Medical Image Analysis
Nov 27, 2024Abstract:Purpose: To explore best-practice approaches for generating synthetic chest X-ray images and augmenting medical imaging datasets to optimize the performance of deep learning models in downstream tasks like classification and segmentation. Materials and Methods: We utilized a latent diffusion model to condition the generation of synthetic chest X-rays on text prompts and/or segmentation masks. We explored methods like using a proxy model and using radiologist feedback to improve the quality of synthetic data. These synthetic images were then generated from relevant disease information or geometrically transformed segmentation masks and added to ground truth training set images from the CheXpert, CANDID-PTX, SIIM, and RSNA Pneumonia datasets to measure improvements in classification and segmentation model performance on the test sets. F1 and Dice scores were used to evaluate classification and segmentation respectively. One-tailed t-tests with Bonferroni correction assessed the statistical significance of performance improvements with synthetic data. Results: Across all experiments, the synthetic data we generated resulted in a maximum mean classification F1 score improvement of 0.150453 (CI: 0.099108-0.201798; P=0.0031) compared to using only real data. For segmentation, the maximum Dice score improvement was 0.14575 (CI: 0.108267-0.183233; P=0.0064). Conclusion: Best practices for generating synthetic chest X-ray images for downstream tasks include conditioning on single-disease labels or geometrically transformed segmentation masks, as well as potentially using proxy modeling for fine-tuning such generations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge