Sophie Ostmeier
MIRIAD: Augmenting LLMs with millions of medical query-response pairs
Jun 06, 2025Abstract:LLMs are bound to transform healthcare with advanced decision support and flexible chat assistants. However, LLMs are prone to generate inaccurate medical content. To ground LLMs in high-quality medical knowledge, LLMs have been equipped with external knowledge via RAG, where unstructured medical knowledge is split into small text chunks that can be selectively retrieved and integrated into the LLMs context. Yet, existing RAG pipelines rely on raw, unstructured medical text, which can be noisy, uncurated and difficult for LLMs to effectively leverage. Systematic approaches to organize medical knowledge to best surface it to LLMs are generally lacking. To address these challenges, we introduce MIRIAD, a large-scale, curated corpus of 5,821,948 medical QA pairs, each rephrased from and grounded in a passage from peer-reviewed medical literature using a semi-automated pipeline combining LLM generation, filtering, grounding, and human annotation. Unlike prior medical corpora, which rely on unstructured text, MIRIAD encapsulates web-scale medical knowledge in an operationalized query-response format, which enables more targeted retrieval. Experiments on challenging medical QA benchmarks show that augmenting LLMs with MIRIAD improves accuracy up to 6.7% compared to unstructured RAG baselines with the same source corpus and with the same amount of retrieved text. Moreover, MIRIAD improved the ability of LLMs to detect medical hallucinations by 22.5 to 37% (increase in F1 score). We further introduce MIRIAD-Atlas, an interactive map of MIRIAD spanning 56 medical disciplines, enabling clinical users to visually explore, search, and refine medical knowledge. MIRIAD promises to unlock a wealth of down-stream applications, including medical information retrievers, enhanced RAG applications, and knowledge-grounded chat interfaces, which ultimately enables more reliable LLM applications in healthcare.
Automated Structured Radiology Report Generation
May 30, 2025Abstract:Automated radiology report generation from chest X-ray (CXR) images has the potential to improve clinical efficiency and reduce radiologists' workload. However, most datasets, including the publicly available MIMIC-CXR and CheXpert Plus, consist entirely of free-form reports, which are inherently variable and unstructured. This variability poses challenges for both generation and evaluation: existing models struggle to produce consistent, clinically meaningful reports, and standard evaluation metrics fail to capture the nuances of radiological interpretation. To address this, we introduce Structured Radiology Report Generation (SRRG), a new task that reformulates free-text radiology reports into a standardized format, ensuring clarity, consistency, and structured clinical reporting. We create a novel dataset by restructuring reports using large language models (LLMs) following strict structured reporting desiderata. Additionally, we introduce SRR-BERT, a fine-grained disease classification model trained on 55 labels, enabling more precise and clinically informed evaluation of structured reports. To assess report quality, we propose F1-SRR-BERT, a metric that leverages SRR-BERT's hierarchical disease taxonomy to bridge the gap between free-text variability and structured clinical reporting. We validate our dataset through a reader study conducted by five board-certified radiologists and extensive benchmarking experiments.
MedVAE: Efficient Automated Interpretation of Medical Images with Large-Scale Generalizable Autoencoders
Feb 20, 2025



Abstract:Medical images are acquired at high resolutions with large fields of view in order to capture fine-grained features necessary for clinical decision-making. Consequently, training deep learning models on medical images can incur large computational costs. In this work, we address the challenge of downsizing medical images in order to improve downstream computational efficiency while preserving clinically-relevant features. We introduce MedVAE, a family of six large-scale 2D and 3D autoencoders capable of encoding medical images as downsized latent representations and decoding latent representations back to high-resolution images. We train MedVAE autoencoders using a novel two-stage training approach with 1,052,730 medical images. Across diverse tasks obtained from 20 medical image datasets, we demonstrate that (1) utilizing MedVAE latent representations in place of high-resolution images when training downstream models can lead to efficiency benefits (up to 70x improvement in throughput) while simultaneously preserving clinically-relevant features and (2) MedVAE can decode latent representations back to high-resolution images with high fidelity. Our work demonstrates that large-scale, generalizable autoencoders can help address critical efficiency challenges in the medical domain. Our code is available at https://github.com/StanfordMIMI/MedVAE.
Preference Fine-Tuning for Factuality in Chest X-Ray Interpretation Models Without Human Feedback
Oct 09, 2024



Abstract:Radiologists play a crucial role by translating medical images into medical reports. However, the field faces staffing shortages and increasing workloads. While automated approaches using vision-language models (VLMs) show promise as assistants, they require exceptionally high accuracy. Most current VLMs in radiology rely solely on supervised fine-tuning (SFT). Meanwhile, in the general domain, additional preference fine-tuning has become standard practice. The challenge in radiology lies in the prohibitive cost of obtaining radiologist feedback. We propose a scalable automated preference alignment technique for VLMs in radiology, focusing on chest X-ray (CXR) report generation. Our method leverages publicly available datasets with an LLM-as-a-Judge mechanism, eliminating the need for additional expert radiologist feedback. We evaluate and benchmark five direct alignment algorithms (DAAs). Our results show up to a 57.4% improvement in average GREEN scores, a LLM-based metric for evaluating CXR reports, and a 9.2% increase in an average across six metrics (domain specific and general), compared to the SFT baseline. We study reward overoptimization via length exploitation, with reports lengthening by up to 3.2x. To assess a potential alignment tax, we benchmark on six additional diverse tasks, finding no significant degradations. A reader study involving four board-certified radiologists indicates win rates of up to 0.62 over the SFT baseline, while significantly penalizing verbosity. Our analysis provides actionable insights for the development of VLMs in high-stakes fields like radiology.
LieRE: Generalizing Rotary Position Encodings
Jun 14, 2024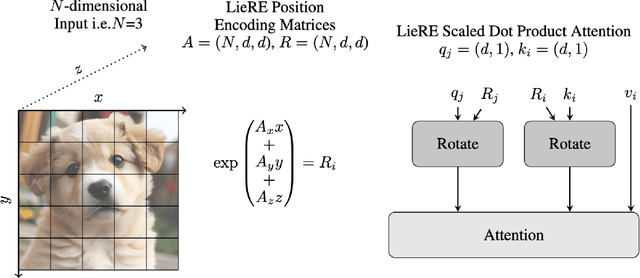
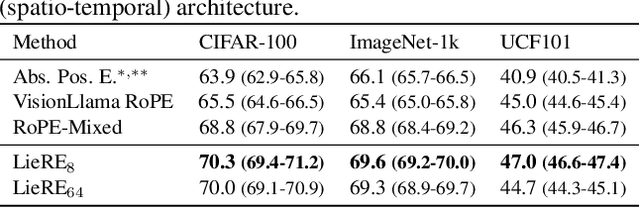
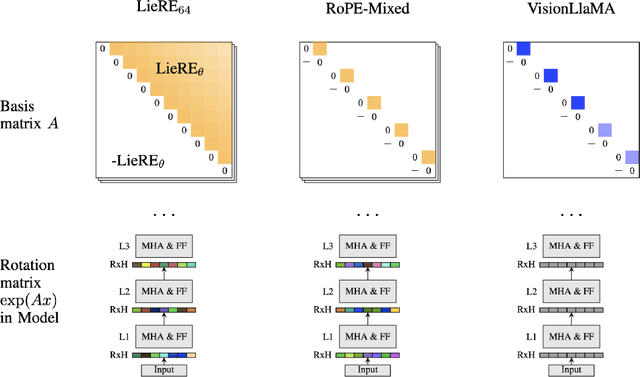
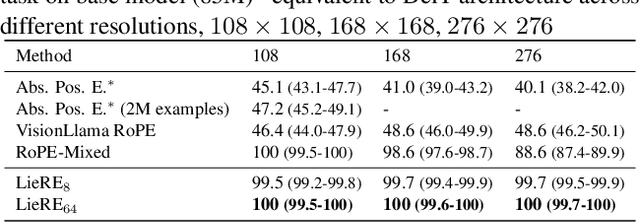
Abstract:While Rotary Position Embeddings (RoPE) for natural language performs well and has become widely adopted, its adoption for other modalities has been slower. Here, we introduce Lie group Relative position Encodings (LieRE) that goes beyond RoPE in supporting higher dimensional inputs. We evaluate the performance of LieRE on 2D and 3D image classification tasks and observe that LieRE leads to marked improvements in performance (up to 6%), training efficiency (3.5x reduction), data efficiency (30%) compared to the baselines of RoFormer, DeiT III, RoPE-Mixed and Vision-Llama
Merlin: A Vision Language Foundation Model for 3D Computed Tomography
Jun 10, 2024



Abstract:Over 85 million computed tomography (CT) scans are performed annually in the US, of which approximately one quarter focus on the abdomen. Given the current radiologist shortage, there is a large impetus to use artificial intelligence to alleviate the burden of interpreting these complex imaging studies. Prior state-of-the-art approaches for automated medical image interpretation leverage vision language models (VLMs). However, current medical VLMs are generally limited to 2D images and short reports, and do not leverage electronic health record (EHR) data for supervision. We introduce Merlin - a 3D VLM that we train using paired CT scans (6+ million images from 15,331 CTs), EHR diagnosis codes (1.8+ million codes), and radiology reports (6+ million tokens). We evaluate Merlin on 6 task types and 752 individual tasks. The non-adapted (off-the-shelf) tasks include zero-shot findings classification (31 findings), phenotype classification (692 phenotypes), and zero-shot cross-modal retrieval (image to findings and image to impressions), while model adapted tasks include 5-year disease prediction (6 diseases), radiology report generation, and 3D semantic segmentation (20 organs). We perform internal validation on a test set of 5,137 CTs, and external validation on 7,000 clinical CTs and on two public CT datasets (VerSe, TotalSegmentator). Beyond these clinically-relevant evaluations, we assess the efficacy of various network architectures and training strategies to depict that Merlin has favorable performance to existing task-specific baselines. We derive data scaling laws to empirically assess training data needs for requisite downstream task performance. Furthermore, unlike conventional VLMs that require hundreds of GPUs for training, we perform all training on a single GPU.
GREEN: Generative Radiology Report Evaluation and Error Notation
May 06, 2024
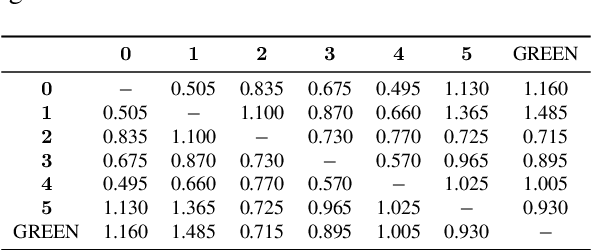


Abstract:Evaluating radiology reports is a challenging problem as factual correctness is extremely important due to the need for accurate medical communication about medical images. Existing automatic evaluation metrics either suffer from failing to consider factual correctness (e.g., BLEU and ROUGE) or are limited in their interpretability (e.g., F1CheXpert and F1RadGraph). In this paper, we introduce GREEN (Generative Radiology Report Evaluation and Error Notation), a radiology report generation metric that leverages the natural language understanding of language models to identify and explain clinically significant errors in candidate reports, both quantitatively and qualitatively. Compared to current metrics, GREEN offers: 1) a score aligned with expert preferences, 2) human interpretable explanations of clinically significant errors, enabling feedback loops with end-users, and 3) a lightweight open-source method that reaches the performance of commercial counterparts. We validate our GREEN metric by comparing it to GPT-4, as well as to error counts of 6 experts and preferences of 2 experts. Our method demonstrates not only higher correlation with expert error counts, but simultaneously higher alignment with expert preferences when compared to previous approaches."
Random Expert Sampling for Deep Learning Segmentation of Acute Ischemic Stroke on Non-contrast CT
Sep 07, 2023Abstract:Purpose: Multi-expert deep learning training methods to automatically quantify ischemic brain tissue on Non-Contrast CT Materials and Methods: The data set consisted of 260 Non-Contrast CTs from 233 patients of acute ischemic stroke patients recruited in the DEFUSE 3 trial. A benchmark U-Net was trained on the reference annotations of three experienced neuroradiologists to segment ischemic brain tissue using majority vote and random expert sampling training schemes. We used a one-sided Wilcoxon signed-rank test on a set of segmentation metrics to compare bootstrapped point estimates of the training schemes with the inter-expert agreement and ratio of variance for consistency analysis. We further compare volumes with the 24h-follow-up DWI (final infarct core) in the patient subgroup with full reperfusion and we test volumes for correlation to the clinical outcome (mRS after 30 and 90 days) with the Spearman method. Results: Random expert sampling leads to a model that shows better agreement with experts than experts agree among themselves and better agreement than the agreement between experts and a majority-vote model performance (Surface Dice at Tolerance 5mm improvement of 61% to 0.70 +- 0.03 and Dice improvement of 25% to 0.50 +- 0.04). The model-based predicted volume similarly estimated the final infarct volume and correlated better to the clinical outcome than CT perfusion. Conclusion: A model trained on random expert sampling can identify the presence and location of acute ischemic brain tissue on Non-Contrast CT similar to CT perfusion and with better consistency than experts. This may further secure the selection of patients eligible for endovascular treatment in less specialized hospitals.
Non-inferiority of Deep Learning Model to Segment Acute Stroke on Non-contrast CT Compared to Neuroradiologists
Nov 24, 2022Abstract:Purpose: To develop a deep learning model to segment the acute ischemic infarct on non-contrast Computed Tomography (NCCT). Materials and Methods In this retrospective study, 227 Head NCCT examinations from 200 patients enrolled in the multicenter DEFUSE 3 trial were included. Three experienced neuroradiologists (experts A, B and C) independently segmented the acute infarct on each study. The dataset was randomly split into 5 folds with training and validation cases. A 3D deep Convolutional Neural Network (CNN) architecture was optimized for the data set properties and task needs. The input to the model was the NCCT and the output was a segmentation mask. The model was trained and optimized on expert A. The outcome was assessed by a set of volume, overlap and distance metrics. The predicted segmentations of the best model and expert A were compared to experts B and C. Then we used a paired Wilcoxon signed-rank test in a one-sided test procedure for all metrics to test for non-inferiority in terms of bias and precision. Results: The best performing model reached a Surface Dice at Tolerance (SDT)5mm of 0.68 \pm 0.04. The predictions were non-inferior when compared to independent experts in terms of bias and precision (paired one-sided test procedure for differences in medians and bootstrapped standard deviations with non-inferior boundaries of -0.05, 2ml, and 2mm, p < 0.05, n=200). Conclusion: For the segmentation of acute ischemic stroke on NCCT, our 3D CNN trained with the annotations of one neuroradiologist is non-inferior when compared to two independent neuroradiologists.
Evaluation of Medical Image Segmentation Models for Uncertain, Small or Empty Reference Annotations
Sep 30, 2022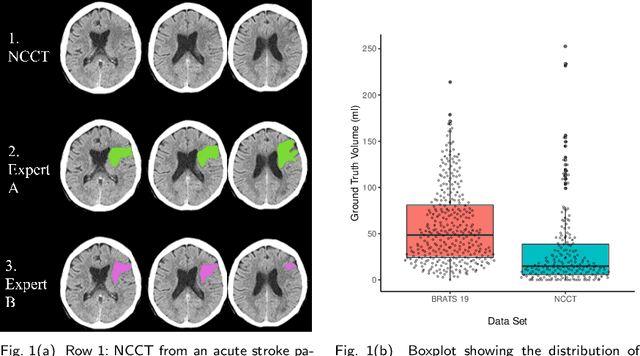

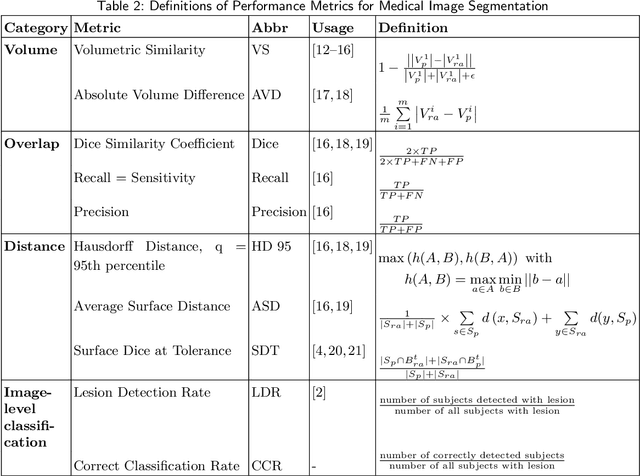
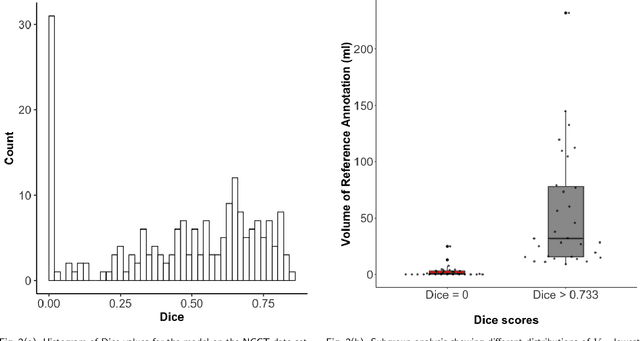
Abstract:Performance metrics for medical image segmentation models are used to measure agreement between the reference annotation and the prediction. A common set of metrics is used in the development of such models to make results more comparable. However, there is a mismatch between the distributions in public data sets and cases encountered in clinical practice. Many common metrics fail to measure the impact of this mismatch, especially for clinical data sets containing uncertain, small or empty reference annotation. Thus, models may not be validated for clinically meaningful agreement by such metrics. Dimensions of evaluating clinical value include independence from reference annotation volume size, consideration of uncertainty of reference annotations, reward of volumetric and/or location agreement and reward of correct classification of empty reference annotations. Unlike common public data sets, our in-house data set is more representative. It contains uncertain, small or empty reference annotations. We examine publicly available metrics on the predictions of a deep learning framework in order to identify for which settings common metrics provide clinical meaningful results. We compare to a public benchmark data set without uncertain, small or empty reference annotations. https://github.com/SophieOstmeier/UncertainSmallEmpty
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge