Jeremy J. Heit
Random Expert Sampling for Deep Learning Segmentation of Acute Ischemic Stroke on Non-contrast CT
Sep 07, 2023Abstract:Purpose: Multi-expert deep learning training methods to automatically quantify ischemic brain tissue on Non-Contrast CT Materials and Methods: The data set consisted of 260 Non-Contrast CTs from 233 patients of acute ischemic stroke patients recruited in the DEFUSE 3 trial. A benchmark U-Net was trained on the reference annotations of three experienced neuroradiologists to segment ischemic brain tissue using majority vote and random expert sampling training schemes. We used a one-sided Wilcoxon signed-rank test on a set of segmentation metrics to compare bootstrapped point estimates of the training schemes with the inter-expert agreement and ratio of variance for consistency analysis. We further compare volumes with the 24h-follow-up DWI (final infarct core) in the patient subgroup with full reperfusion and we test volumes for correlation to the clinical outcome (mRS after 30 and 90 days) with the Spearman method. Results: Random expert sampling leads to a model that shows better agreement with experts than experts agree among themselves and better agreement than the agreement between experts and a majority-vote model performance (Surface Dice at Tolerance 5mm improvement of 61% to 0.70 +- 0.03 and Dice improvement of 25% to 0.50 +- 0.04). The model-based predicted volume similarly estimated the final infarct volume and correlated better to the clinical outcome than CT perfusion. Conclusion: A model trained on random expert sampling can identify the presence and location of acute ischemic brain tissue on Non-Contrast CT similar to CT perfusion and with better consistency than experts. This may further secure the selection of patients eligible for endovascular treatment in less specialized hospitals.
Non-inferiority of Deep Learning Model to Segment Acute Stroke on Non-contrast CT Compared to Neuroradiologists
Nov 24, 2022Abstract:Purpose: To develop a deep learning model to segment the acute ischemic infarct on non-contrast Computed Tomography (NCCT). Materials and Methods In this retrospective study, 227 Head NCCT examinations from 200 patients enrolled in the multicenter DEFUSE 3 trial were included. Three experienced neuroradiologists (experts A, B and C) independently segmented the acute infarct on each study. The dataset was randomly split into 5 folds with training and validation cases. A 3D deep Convolutional Neural Network (CNN) architecture was optimized for the data set properties and task needs. The input to the model was the NCCT and the output was a segmentation mask. The model was trained and optimized on expert A. The outcome was assessed by a set of volume, overlap and distance metrics. The predicted segmentations of the best model and expert A were compared to experts B and C. Then we used a paired Wilcoxon signed-rank test in a one-sided test procedure for all metrics to test for non-inferiority in terms of bias and precision. Results: The best performing model reached a Surface Dice at Tolerance (SDT)5mm of 0.68 \pm 0.04. The predictions were non-inferior when compared to independent experts in terms of bias and precision (paired one-sided test procedure for differences in medians and bootstrapped standard deviations with non-inferior boundaries of -0.05, 2ml, and 2mm, p < 0.05, n=200). Conclusion: For the segmentation of acute ischemic stroke on NCCT, our 3D CNN trained with the annotations of one neuroradiologist is non-inferior when compared to two independent neuroradiologists.
Evaluation of Medical Image Segmentation Models for Uncertain, Small or Empty Reference Annotations
Sep 30, 2022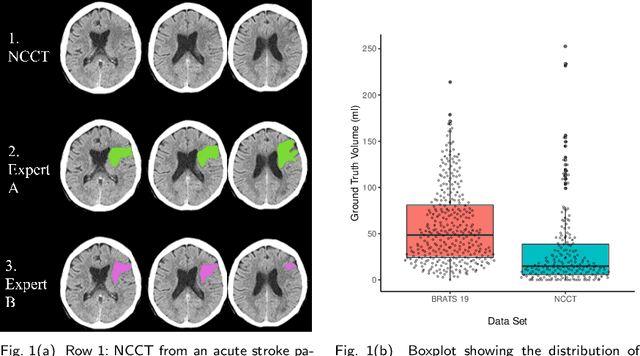

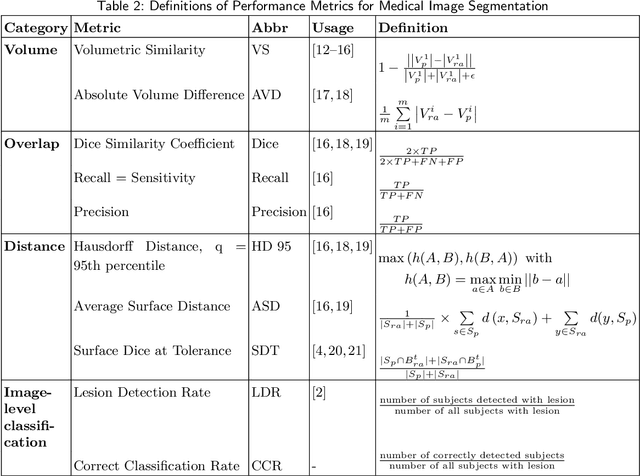
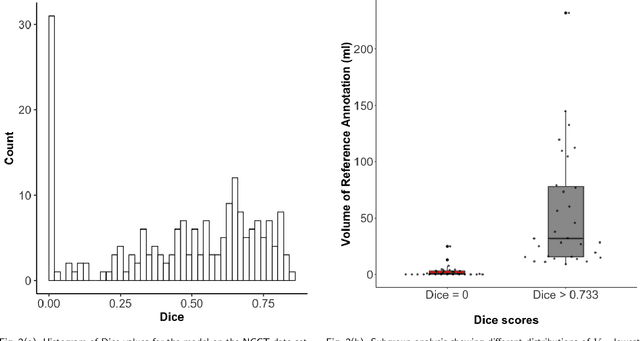
Abstract:Performance metrics for medical image segmentation models are used to measure agreement between the reference annotation and the prediction. A common set of metrics is used in the development of such models to make results more comparable. However, there is a mismatch between the distributions in public data sets and cases encountered in clinical practice. Many common metrics fail to measure the impact of this mismatch, especially for clinical data sets containing uncertain, small or empty reference annotation. Thus, models may not be validated for clinically meaningful agreement by such metrics. Dimensions of evaluating clinical value include independence from reference annotation volume size, consideration of uncertainty of reference annotations, reward of volumetric and/or location agreement and reward of correct classification of empty reference annotations. Unlike common public data sets, our in-house data set is more representative. It contains uncertain, small or empty reference annotations. We examine publicly available metrics on the predictions of a deep learning framework in order to identify for which settings common metrics provide clinical meaningful results. We compare to a public benchmark data set without uncertain, small or empty reference annotations. https://github.com/SophieOstmeier/UncertainSmallEmpty
Diffusion-Weighted Magnetic Resonance Brain Images Generation with Generative Adversarial Networks and Variational Autoencoders: A Comparison Study
Jun 24, 2020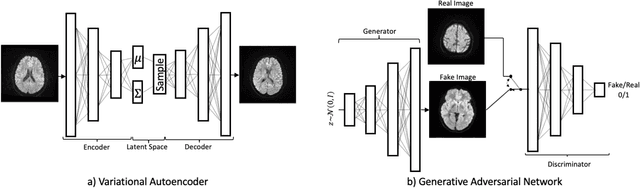
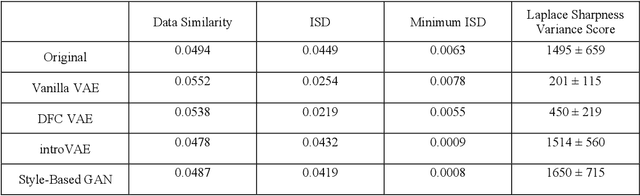
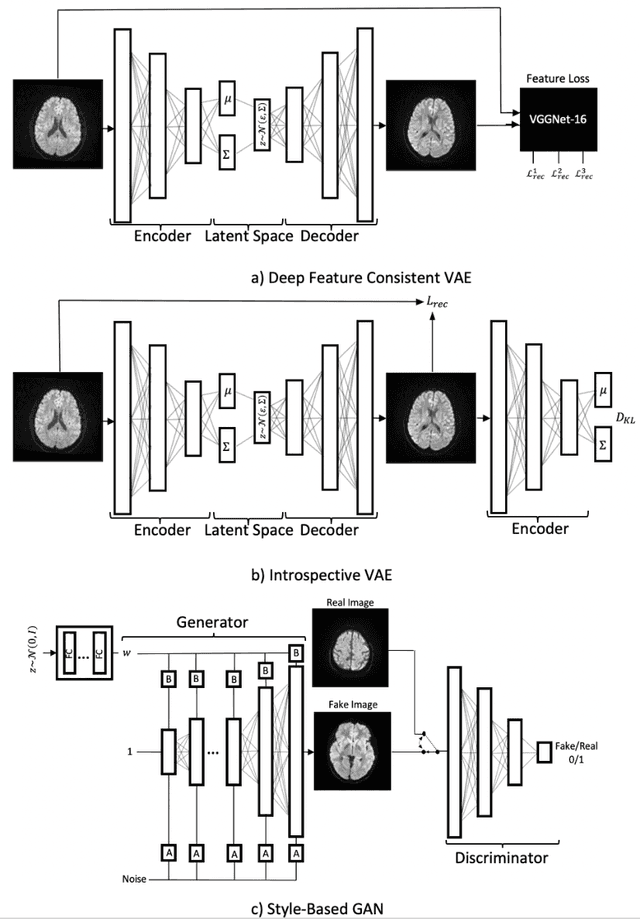
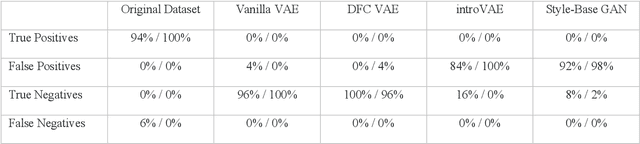
Abstract:We show that high quality, diverse and realistic-looking diffusion-weighted magnetic resonance images can be synthesized using deep generative models. Based on professional neuroradiologists' evaluations and diverse metrics with respect to quality and diversity of the generated synthetic brain images, we present two networks, the Introspective Variational Autoencoder and the Style-Based GAN, that qualify for data augmentation in the medical field, where information is saved in a dispatched and inhomogeneous way and access to it is in many aspects restricted.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge