Greg Zaharchuk
for the Alzheimer's Disease Neuroimaging Initiative
Score-based Generative Diffusion Models to Synthesize Full-dose FDG Brain PET from MRI in Epilepsy Patients
Jun 12, 2025Abstract:Fluorodeoxyglucose (FDG) PET to evaluate patients with epilepsy is one of the most common applications for simultaneous PET/MRI, given the need to image both brain structure and metabolism, but is suboptimal due to the radiation dose in this young population. Little work has been done synthesizing diagnostic quality PET images from MRI data or MRI data with ultralow-dose PET using advanced generative AI methods, such as diffusion models, with attention to clinical evaluations tailored for the epilepsy population. Here we compared the performance of diffusion- and non-diffusion-based deep learning models for the MRI-to-PET image translation task for epilepsy imaging using simultaneous PET/MRI in 52 subjects (40 train/2 validate/10 hold-out test). We tested three different models: 2 score-based generative diffusion models (SGM-Karras Diffusion [SGM-KD] and SGM-variance preserving [SGM-VP]) and a Transformer-Unet. We report results on standard image processing metrics as well as clinically relevant metrics, including congruency measures (Congruence Index and Congruency Mean Absolute Error) that assess hemispheric metabolic asymmetry, which is a key part of the clinical analysis of these images. The SGM-KD produced the best qualitative and quantitative results when synthesizing PET purely from T1w and T2 FLAIR images with the least mean absolute error in whole-brain specific uptake value ratio (SUVR) and highest intraclass correlation coefficient. When 1% low-dose PET images are included in the inputs, all models improve significantly and are interchangeable for quantitative performance and visual quality. In summary, SGMs hold great potential for pure MRI-to-PET translation, while all 3 model types can synthesize full-dose FDG-PET accurately using MRI and ultralow-dose PET.
Deep Learning-Based Prediction of PET Amyloid Status Using Multi-Contrast MRI
Nov 18, 2024



Abstract:Identifying amyloid-beta positive patients is crucial for determining eligibility for Alzheimer's disease (AD) clinical trials and new disease-modifying treatments, but currently requires PET or CSF sampling. Previous MRI-based deep learning models for predicting amyloid positivity, using only T1w sequences, have shown moderate performance. We trained deep learning models to predict amyloid PET positivity and evaluated whether multi-contrast inputs improve performance. A total of 4,058 exams with multi-contrast MRI and PET-based quantitative amyloid deposition were obtained from three public datasets: the Alzheimer's Disease Neuroimaging Initiative (ADNI), the Open Access Series of Imaging Studies 3 (OASIS3), and the Anti-Amyloid Treatment in Asymptomatic Alzheimer's Disease (A4). Two separate EfficientNet models were trained for amyloid positivity prediction: one with only T1w images and the other with both T1w and T2-FLAIR images as network inputs. The area under the curve (AUC), accuracy, sensitivity, and specificity were determined using an internal held-out test set. The trained models were further evaluated using an external test set. In the held-out test sets, the T1w and T1w+T2FLAIR models demonstrated AUCs of 0.62 (95% CI: 0.60, 0.64) and 0.67 (95% CI: 0.64, 0.70) (p = 0.006); accuracies were 61% (95% CI: 60%, 63%) and 64% (95% CI: 62%, 66%) (p = 0.008); sensitivities were 0.88 and 0.71; and specificities were 0.23 and 0.53, respectively. The trained models showed similar performance in the external test set. Performance of the current model on both test sets exceeded that of the publicly available model. In conclusion, the use of multi-contrast MRI, specifically incorporating T2-FLAIR in addition to T1w images, significantly improved the predictive accuracy of PET-determined amyloid status from MRI scans using a deep learning approach.
LSOR: Longitudinally-Consistent Self-Organized Representation Learning
Sep 30, 2023Abstract:Interpretability is a key issue when applying deep learning models to longitudinal brain MRIs. One way to address this issue is by visualizing the high-dimensional latent spaces generated by deep learning via self-organizing maps (SOM). SOM separates the latent space into clusters and then maps the cluster centers to a discrete (typically 2D) grid preserving the high-dimensional relationship between clusters. However, learning SOM in a high-dimensional latent space tends to be unstable, especially in a self-supervision setting. Furthermore, the learned SOM grid does not necessarily capture clinically interesting information, such as brain age. To resolve these issues, we propose the first self-supervised SOM approach that derives a high-dimensional, interpretable representation stratified by brain age solely based on longitudinal brain MRIs (i.e., without demographic or cognitive information). Called Longitudinally-consistent Self-Organized Representation learning (LSOR), the method is stable during training as it relies on soft clustering (vs. the hard cluster assignments used by existing SOM). Furthermore, our approach generates a latent space stratified according to brain age by aligning trajectories inferred from longitudinal MRIs to the reference vector associated with the corresponding SOM cluster. When applied to longitudinal MRIs of the Alzheimer's Disease Neuroimaging Initiative (ADNI, N=632), LSOR generates an interpretable latent space and achieves comparable or higher accuracy than the state-of-the-art representations with respect to the downstream tasks of classification (static vs. progressive mild cognitive impairment) and regression (determining ADAS-Cog score of all subjects). The code is available at https://github.com/ouyangjiahong/longitudinal-som-single-modality.
Random Expert Sampling for Deep Learning Segmentation of Acute Ischemic Stroke on Non-contrast CT
Sep 07, 2023Abstract:Purpose: Multi-expert deep learning training methods to automatically quantify ischemic brain tissue on Non-Contrast CT Materials and Methods: The data set consisted of 260 Non-Contrast CTs from 233 patients of acute ischemic stroke patients recruited in the DEFUSE 3 trial. A benchmark U-Net was trained on the reference annotations of three experienced neuroradiologists to segment ischemic brain tissue using majority vote and random expert sampling training schemes. We used a one-sided Wilcoxon signed-rank test on a set of segmentation metrics to compare bootstrapped point estimates of the training schemes with the inter-expert agreement and ratio of variance for consistency analysis. We further compare volumes with the 24h-follow-up DWI (final infarct core) in the patient subgroup with full reperfusion and we test volumes for correlation to the clinical outcome (mRS after 30 and 90 days) with the Spearman method. Results: Random expert sampling leads to a model that shows better agreement with experts than experts agree among themselves and better agreement than the agreement between experts and a majority-vote model performance (Surface Dice at Tolerance 5mm improvement of 61% to 0.70 +- 0.03 and Dice improvement of 25% to 0.50 +- 0.04). The model-based predicted volume similarly estimated the final infarct volume and correlated better to the clinical outcome than CT perfusion. Conclusion: A model trained on random expert sampling can identify the presence and location of acute ischemic brain tissue on Non-Contrast CT similar to CT perfusion and with better consistency than experts. This may further secure the selection of patients eligible for endovascular treatment in less specialized hospitals.
Simulation of Arbitrary Level Contrast Dose in MRI Using an Iterative Global Transformer Model
Jul 22, 2023



Abstract:Deep learning (DL) based contrast dose reduction and elimination in MRI imaging is gaining traction, given the detrimental effects of Gadolinium-based Contrast Agents (GBCAs). These DL algorithms are however limited by the availability of high quality low dose datasets. Additionally, different types of GBCAs and pathologies require different dose levels for the DL algorithms to work reliably. In this work, we formulate a novel transformer (Gformer) based iterative modelling approach for the synthesis of images with arbitrary contrast enhancement that corresponds to different dose levels. The proposed Gformer incorporates a sub-sampling based attention mechanism and a rotational shift module that captures the various contrast related features. Quantitative evaluation indicates that the proposed model performs better than other state-of-the-art methods. We further perform quantitative evaluation on downstream tasks such as dose reduction and tumor segmentation to demonstrate the clinical utility.
Non-inferiority of Deep Learning Model to Segment Acute Stroke on Non-contrast CT Compared to Neuroradiologists
Nov 24, 2022Abstract:Purpose: To develop a deep learning model to segment the acute ischemic infarct on non-contrast Computed Tomography (NCCT). Materials and Methods In this retrospective study, 227 Head NCCT examinations from 200 patients enrolled in the multicenter DEFUSE 3 trial were included. Three experienced neuroradiologists (experts A, B and C) independently segmented the acute infarct on each study. The dataset was randomly split into 5 folds with training and validation cases. A 3D deep Convolutional Neural Network (CNN) architecture was optimized for the data set properties and task needs. The input to the model was the NCCT and the output was a segmentation mask. The model was trained and optimized on expert A. The outcome was assessed by a set of volume, overlap and distance metrics. The predicted segmentations of the best model and expert A were compared to experts B and C. Then we used a paired Wilcoxon signed-rank test in a one-sided test procedure for all metrics to test for non-inferiority in terms of bias and precision. Results: The best performing model reached a Surface Dice at Tolerance (SDT)5mm of 0.68 \pm 0.04. The predictions were non-inferior when compared to independent experts in terms of bias and precision (paired one-sided test procedure for differences in medians and bootstrapped standard deviations with non-inferior boundaries of -0.05, 2ml, and 2mm, p < 0.05, n=200). Conclusion: For the segmentation of acute ischemic stroke on NCCT, our 3D CNN trained with the annotations of one neuroradiologist is non-inferior when compared to two independent neuroradiologists.
Brain MRI-to-PET Synthesis using 3D Convolutional Attention Networks
Nov 22, 2022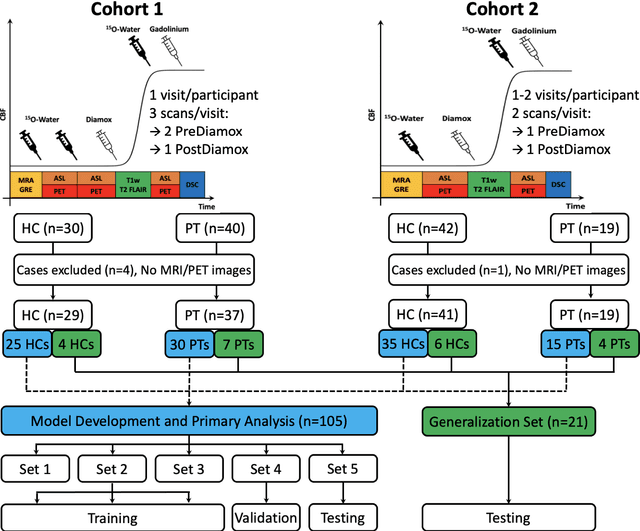
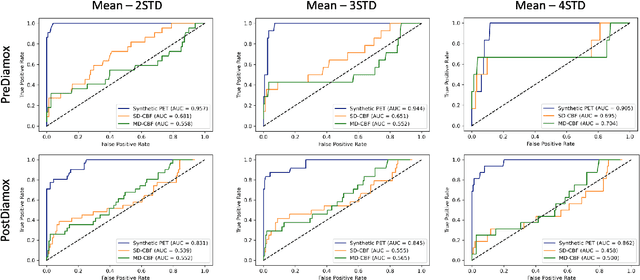
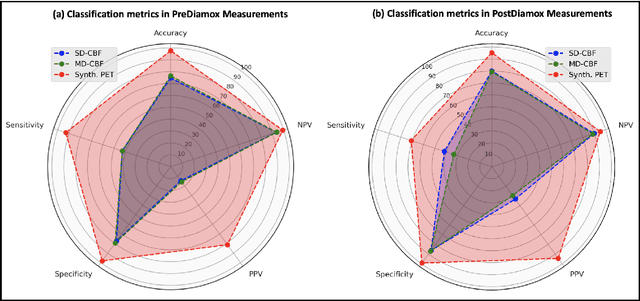
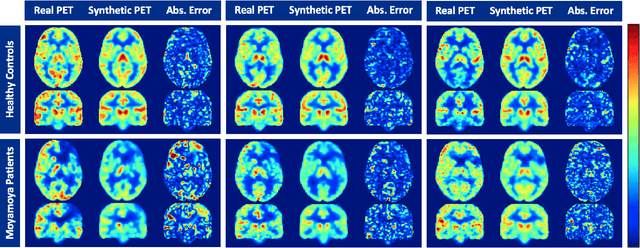
Abstract:Accurate quantification of cerebral blood flow (CBF) is essential for the diagnosis and assessment of a wide range of neurological diseases. Positron emission tomography (PET) with radiolabeled water (15O-water) is considered the gold-standard for the measurement of CBF in humans. PET imaging, however, is not widely available because of its prohibitive costs and use of short-lived radiopharmaceutical tracers that typically require onsite cyclotron production. Magnetic resonance imaging (MRI), in contrast, is more readily accessible and does not involve ionizing radiation. This study presents a convolutional encoder-decoder network with attention mechanisms to predict gold-standard 15O-water PET CBF from multi-sequence MRI scans, thereby eliminating the need for radioactive tracers. Inputs to the prediction model include several commonly used MRI sequences (T1-weighted, T2-FLAIR, and arterial spin labeling). The model was trained and validated using 5-fold cross-validation in a group of 126 subjects consisting of healthy controls and cerebrovascular disease patients, all of whom underwent simultaneous $15O-water PET/MRI. The results show that such a model can successfully synthesize high-quality PET CBF measurements (with an average SSIM of 0.924 and PSNR of 38.8 dB) and is more accurate compared to concurrent and previous PET synthesis methods. We also demonstrate the clinical significance of the proposed algorithm by evaluating the agreement for identifying the vascular territories with abnormally low CBF. Such methods may enable more widespread and accurate CBF evaluation in larger cohorts who cannot undergo PET imaging due to radiation concerns, lack of access, or logistic challenges.
One Model to Synthesize Them All: Multi-contrast Multi-scale Transformer for Missing Data Imputation
Apr 28, 2022


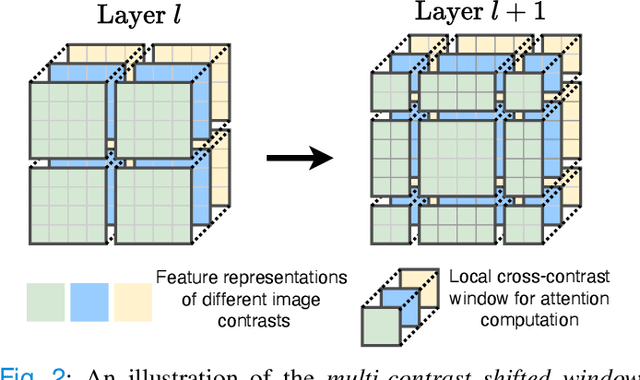
Abstract:Multi-contrast magnetic resonance imaging (MRI) is widely used in clinical practice as each contrast provides complementary information. However, the availability of each contrast may vary amongst patients in reality. This poses challenges to both radiologists and automated image analysis algorithms. A general approach for tackling this problem is missing data imputation, which aims to synthesize the missing contrasts from existing ones. While several convolutional neural network (CNN) based algorithms have been proposed, they suffer from the fundamental limitations of CNN models, such as requirement for fixed numbers of input and output channels, inability to capture long-range dependencies, and lack of interpretability. In this paper, we formulate missing data imputation as a sequence-to-sequence learning problem and propose a multi-contrast multi-scale Transformer (MMT), which can take any subset of input contrasts and synthesize those that are missing. MMT consists of a multi-scale Transformer encoder that builds hierarchical representations of inputs combined with a multi-scale Transformer decoder that generates the outputs in a coarse-to-fine fashion. Thanks to the proposed multi-contrast Swin Transformer blocks, it can efficiently capture intra- and inter-contrast dependencies for accurate image synthesis. Moreover, MMT is inherently interpretable. It allows us to understand the importance of each input contrast in different regions by analyzing the in-built attention maps of Transformer blocks in the decoder. Extensive experiments on two large-scale multi-contrast MRI datasets demonstrate that MMT outperforms the state-of-the-art methods quantitatively and qualitatively.
Multi-task Deep Learning for Cerebrovascular Disease Classification and MRI-to-PET Translation
Feb 12, 2022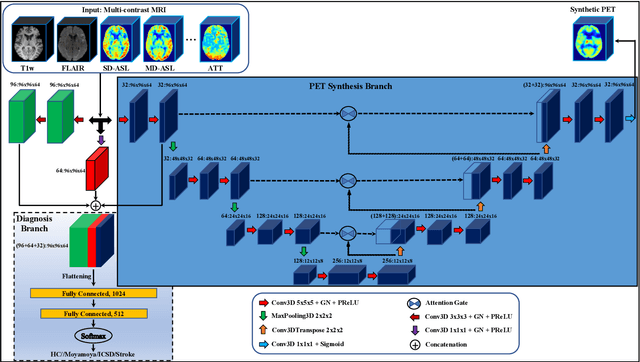
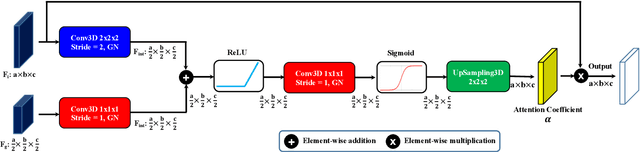
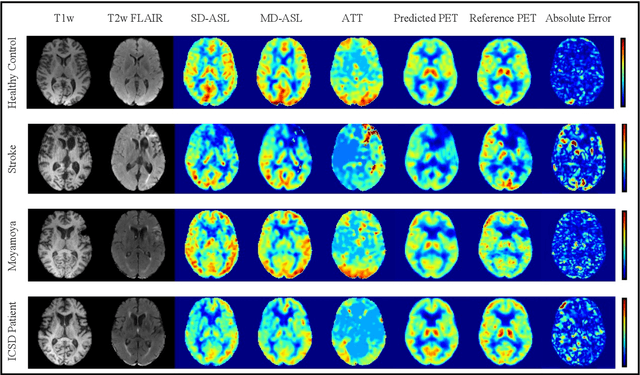
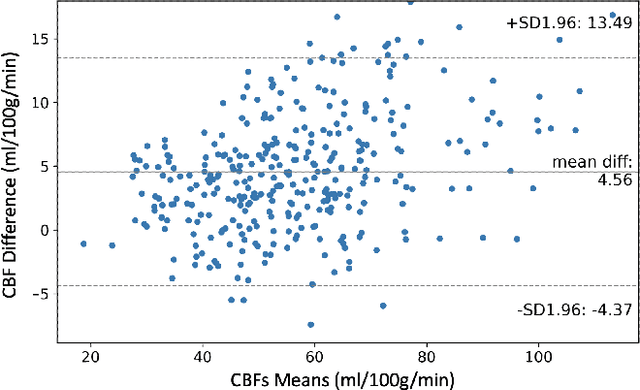
Abstract:Accurate quantification of cerebral blood flow (CBF) is essential for the diagnosis and assessment of cerebrovascular diseases such as Moyamoya, carotid stenosis, aneurysms, and stroke. Positron emission tomography (PET) is currently regarded as the gold standard for the measurement of CBF in the human brain. PET imaging, however, is not widely available because of its prohibitive costs, use of ionizing radiation, and logistical challenges, which require a co-localized cyclotron to deliver the 2 min half-life Oxygen-15 radioisotope. Magnetic resonance imaging (MRI), in contrast, is more readily available and does not involve ionizing radiation. In this study, we propose a multi-task learning framework for brain MRI-to-PET translation and disease diagnosis. The proposed framework comprises two prime networks: (1) an attention-based 3D encoder-decoder convolutional neural network (CNN) that synthesizes high-quality PET CBF maps from multi-contrast MRI images, and (2) a multi-scale 3D CNN that identifies the brain disease corresponding to the input MRI images. Our multi-task framework yields promising results on the task of MRI-to-PET translation, achieving an average structural similarity index (SSIM) of 0.94 and peak signal-to-noise ratio (PSNR) of 38dB on a cohort of 120 subjects. In addition, we show that integrating multiple MRI modalities can improve the clinical diagnosis of brain diseases.
Self-Supervised Longitudinal Neighbourhood Embedding
Mar 09, 2021



Abstract:Longitudinal MRIs are often used to capture the gradual deterioration of brain structure and function caused by aging or neurological diseases. Analyzing this data via machine learning generally requires a large number of ground-truth labels, which are often missing or expensive to obtain. Reducing the need for labels, we propose a self-supervised strategy for representation learning named Longitudinal Neighborhood Embedding (LNE). Motivated by concepts in contrastive learning, LNE explicitly models the similarity between trajectory vectors across different subjects. We do so by building a graph in each training iteration defining neighborhoods in the latent space so that the progression direction of a subject follows the direction of its neighbors. This results in a smooth trajectory field that captures the global morphological change of the brain while maintaining the local continuity. We apply LNE to longitudinal T1w MRIs of two neuroimaging studies: a dataset composed of 274 healthy subjects, and Alzheimer's Disease Neuroimaging Initiative (ADNI, N=632). The visualization of the smooth trajectory vector field and superior performance on downstream tasks demonstrate the strength of the proposed method over existing self-supervised methods in extracting information associated with normal aging and in revealing the impact of neurodegenerative disorders. The code is available at \url{https://github.com/ouyangjiahong/longitudinal-neighbourhood-embedding.git}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge