Enhao Gong
One Model to Synthesize Them All: Multi-contrast Multi-scale Transformer for Missing Data Imputation
Apr 28, 2022


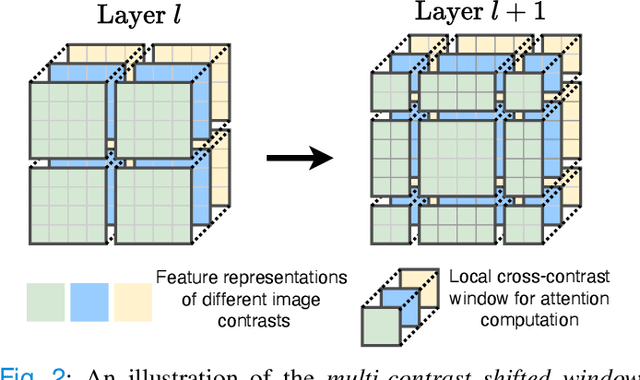
Abstract:Multi-contrast magnetic resonance imaging (MRI) is widely used in clinical practice as each contrast provides complementary information. However, the availability of each contrast may vary amongst patients in reality. This poses challenges to both radiologists and automated image analysis algorithms. A general approach for tackling this problem is missing data imputation, which aims to synthesize the missing contrasts from existing ones. While several convolutional neural network (CNN) based algorithms have been proposed, they suffer from the fundamental limitations of CNN models, such as requirement for fixed numbers of input and output channels, inability to capture long-range dependencies, and lack of interpretability. In this paper, we formulate missing data imputation as a sequence-to-sequence learning problem and propose a multi-contrast multi-scale Transformer (MMT), which can take any subset of input contrasts and synthesize those that are missing. MMT consists of a multi-scale Transformer encoder that builds hierarchical representations of inputs combined with a multi-scale Transformer decoder that generates the outputs in a coarse-to-fine fashion. Thanks to the proposed multi-contrast Swin Transformer blocks, it can efficiently capture intra- and inter-contrast dependencies for accurate image synthesis. Moreover, MMT is inherently interpretable. It allows us to understand the importance of each input contrast in different regions by analyzing the in-built attention maps of Transformer blocks in the decoder. Extensive experiments on two large-scale multi-contrast MRI datasets demonstrate that MMT outperforms the state-of-the-art methods quantitatively and qualitatively.
OUTCOMES: Rapid Under-sampling Optimization achieves up to 50% improvements in reconstruction accuracy for multi-contrast MRI sequences
Mar 08, 2021



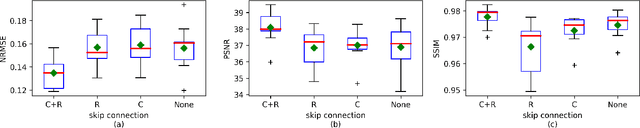
Abstract:Multi-contrast Magnetic Resonance Imaging (MRI) acquisitions from a single scan have tremendous potential to streamline exams and reduce imaging time. However, maintaining clinically feasible scan time necessitates significant undersampling, pushing the limits on compressed sensing and other low-dimensional techniques. During MRI scanning, one of the possible solutions is by using undersampling designs which can effectively improve the acquisition and achieve higher reconstruction accuracy. However, existing undersampling optimization methods are time-consuming and the limited performance prevents their clinical applications. In this paper, we proposed an improved undersampling trajectory optimization scheme to generate an optimized trajectory within seconds and apply it to subsequent multi-contrast MRI datasets on a per-subject basis, where we named it OUTCOMES. By using a data-driven method combined with improved algorithm design, GPU acceleration, and more efficient computation, the proposed method can optimize a trajectory within 5-10 seconds and achieve 30%-50% reconstruction improvement with the same acquisition cost, which makes real-time under-sampling optimization possible for clinical applications.
200x Low-dose PET Reconstruction using Deep Learning
Dec 12, 2017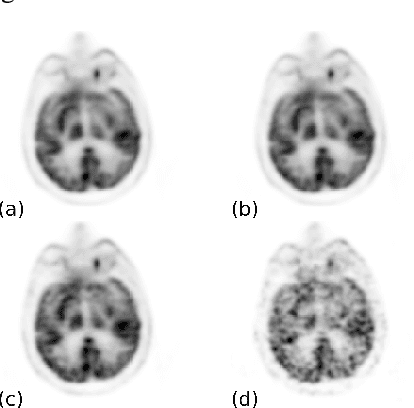
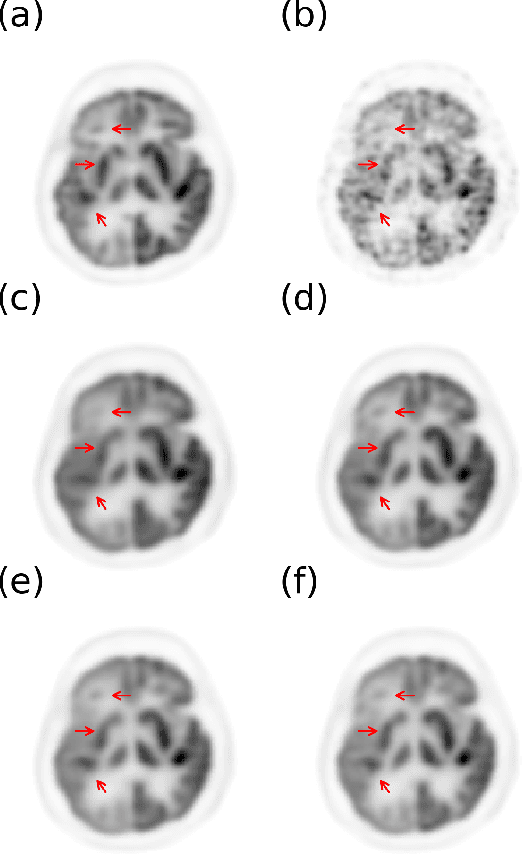
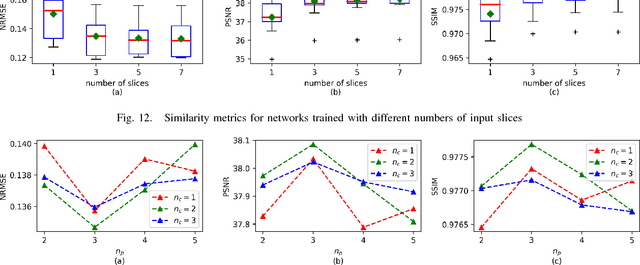
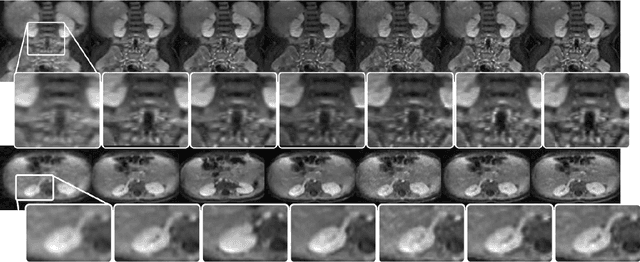
Abstract:Positron emission tomography (PET) is widely used in various clinical applications, including cancer diagnosis, heart disease and neuro disorders. The use of radioactive tracer in PET imaging raises concerns due to the risk of radiation exposure. To minimize this potential risk in PET imaging, efforts have been made to reduce the amount of radio-tracer usage. However, lowing dose results in low Signal-to-Noise-Ratio (SNR) and loss of information, both of which will heavily affect clinical diagnosis. Besides, the ill-conditioning of low-dose PET image reconstruction makes it a difficult problem for iterative reconstruction algorithms. Previous methods proposed are typically complicated and slow, yet still cannot yield satisfactory results at significantly low dose. Here, we propose a deep learning method to resolve this issue with an encoder-decoder residual deep network with concatenate skip connections. Experiments shows the proposed method can reconstruct low-dose PET image to a standard-dose quality with only two-hundredth dose. Different cost functions for training model are explored. Multi-slice input strategy is introduced to provide the network with more structural information and make it more robust to noise. Evaluation on ultra-low-dose clinical data shows that the proposed method can achieve better result than the state-of-the-art methods and reconstruct images with comparable quality using only 0.5% of the original regular dose.
Deep Generative Adversarial Networks for Compressed Sensing Automates MRI
May 31, 2017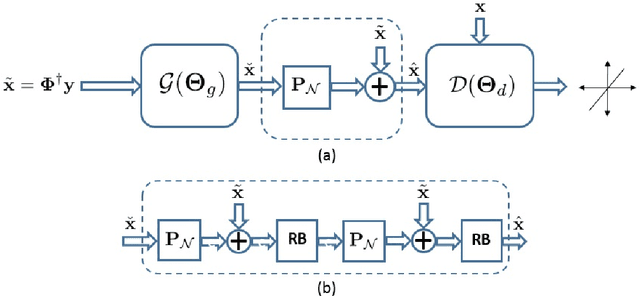
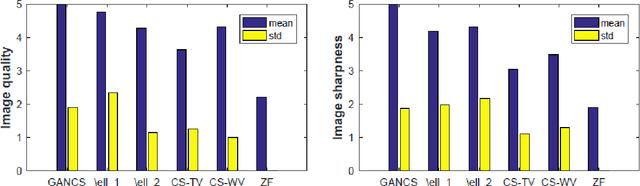
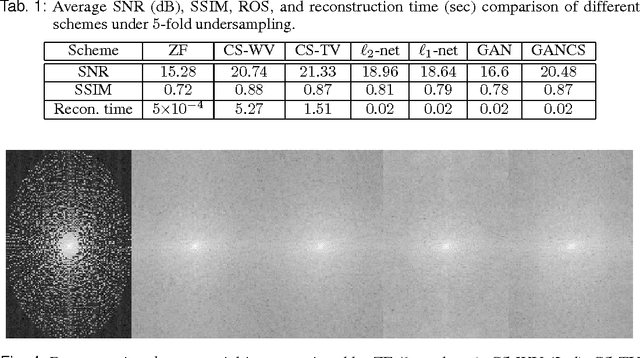
Abstract:Magnetic resonance image (MRI) reconstruction is a severely ill-posed linear inverse task demanding time and resource intensive computations that can substantially trade off {\it accuracy} for {\it speed} in real-time imaging. In addition, state-of-the-art compressed sensing (CS) analytics are not cognizant of the image {\it diagnostic quality}. To cope with these challenges we put forth a novel CS framework that permeates benefits from generative adversarial networks (GAN) to train a (low-dimensional) manifold of diagnostic-quality MR images from historical patients. Leveraging a mixture of least-squares (LS) GANs and pixel-wise $\ell_1$ cost, a deep residual network with skip connections is trained as the generator that learns to remove the {\it aliasing} artifacts by projecting onto the manifold. LSGAN learns the texture details, while $\ell_1$ controls the high-frequency noise. A multilayer convolutional neural network is then jointly trained based on diagnostic quality images to discriminate the projection quality. The test phase performs feed-forward propagation over the generator network that demands a very low computational overhead. Extensive evaluations are performed on a large contrast-enhanced MR dataset of pediatric patients. In particular, images rated based on expert radiologists corroborate that GANCS retrieves high contrast images with detailed texture relative to conventional CS, and pixel-wise schemes. In addition, it offers reconstruction under a few milliseconds, two orders of magnitude faster than state-of-the-art CS-MRI schemes.
DSD: Dense-Sparse-Dense Training for Deep Neural Networks
Feb 21, 2017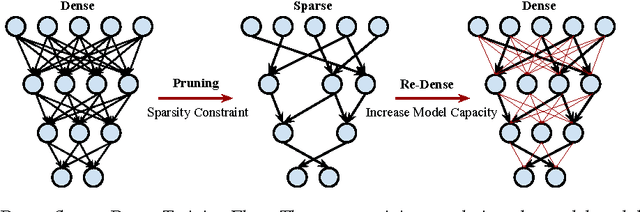
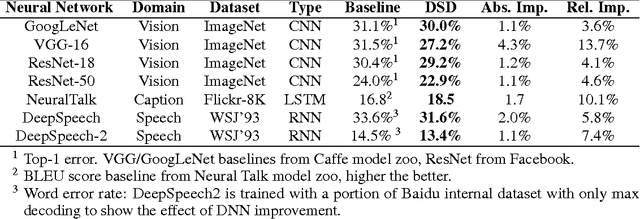
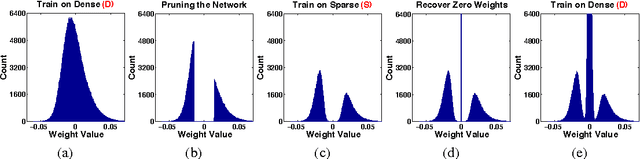
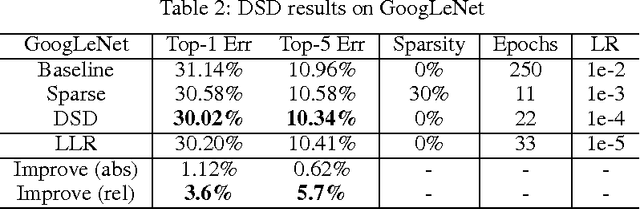
Abstract:Modern deep neural networks have a large number of parameters, making them very hard to train. We propose DSD, a dense-sparse-dense training flow, for regularizing deep neural networks and achieving better optimization performance. In the first D (Dense) step, we train a dense network to learn connection weights and importance. In the S (Sparse) step, we regularize the network by pruning the unimportant connections with small weights and retraining the network given the sparsity constraint. In the final D (re-Dense) step, we increase the model capacity by removing the sparsity constraint, re-initialize the pruned parameters from zero and retrain the whole dense network. Experiments show that DSD training can improve the performance for a wide range of CNNs, RNNs and LSTMs on the tasks of image classification, caption generation and speech recognition. On ImageNet, DSD improved the Top1 accuracy of GoogLeNet by 1.1%, VGG-16 by 4.3%, ResNet-18 by 1.2% and ResNet-50 by 1.1%, respectively. On the WSJ'93 dataset, DSD improved DeepSpeech and DeepSpeech2 WER by 2.0% and 1.1%. On the Flickr-8K dataset, DSD improved the NeuralTalk BLEU score by over 1.7. DSD is easy to use in practice: at training time, DSD incurs only one extra hyper-parameter: the sparsity ratio in the S step. At testing time, DSD doesn't change the network architecture or incur any inference overhead. The consistent and significant performance gain of DSD experiments shows the inadequacy of the current training methods for finding the best local optimum, while DSD effectively achieves superior optimization performance for finding a better solution. DSD models are available to download at https://songhan.github.io/DSD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge