John M. Pauly
OPTIKS: Optimized Gradient Properties Through Timing in K-Space
May 11, 2025



Abstract:A customizable method (OPTIKS) for designing fast trajectory-constrained gradient waveforms with optimized time domain properties was developed. Given a specified multidimensional k-space trajectory, the method optimizes traversal speed (and therefore timing) with position along the trajectory. OPTIKS facilitates optimization of objectives dependent on the time domain gradient waveform and the arc-length domain k-space speed. OPTIKS is applied to design waveforms which limit peripheral nerve stimulation (PNS), minimize mechanical resonance excitation, and reduce acoustic noise. A variety of trajectory examples are presented including spirals, circular echo-planar-imaging, and rosettes. Design performance is evaluated based on duration, standardized PNS models, field measurements, gradient coil back-EMF measurements, and calibrated acoustic measurements. We show reductions in back-EMF of up to 94% and field oscillations up to 91.1%, acoustic noise decreases of up to 9.22 dB, and with efficient use of PNS models speed increases of up to 11.4%. The design method implementation is made available as an open source Python package through GitHub.
AutoSamp: Autoencoding MRI Sampling via Variational Information Maximization
Jun 07, 2023Abstract:Accelerated MRI protocols routinely involve a predefined sampling pattern that undersamples the k-space. Finding an optimal pattern can enhance the reconstruction quality, however this optimization is a challenging task. To address this challenge, we introduce a novel deep learning framework, AutoSamp, based on variational information maximization that enables joint optimization of sampling pattern and reconstruction of MRI scans. We represent the encoder as a non-uniform Fast Fourier Transform that allows continuous optimization of k-space sample locations on a non-Cartesian plane, and the decoder as a deep reconstruction network. Experiments on public MRI datasets show improved reconstruction quality of the proposed AutoSamp method over the prevailing variable density and variable density Poisson disc sampling. We demonstrate that our data-driven sampling optimization method achieves 4.4dB, 2.0dB, 0.75dB, 0.7dB PSNR improvements over reconstruction with Poisson Disc masks for acceleration factors of R = 5, 10, 15, 25, respectively. Furthermore, we analyze the characteristics of the learned sampling patterns with respect to changes in acceleration factor, measurement noise, underlying anatomy, and coil sensitivities. We show that all these factors contribute to the optimization result by affecting the sampling density, k-space coverage and point spread functions of the learned sampling patterns.
Automated MRI Field of View Prescription from Region of Interest Prediction by Intra-stack Attention Neural Network
Nov 09, 2022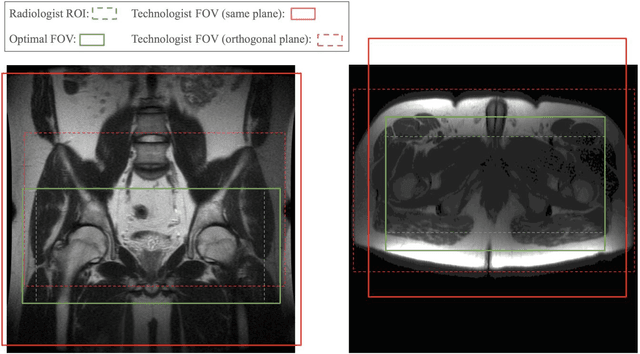
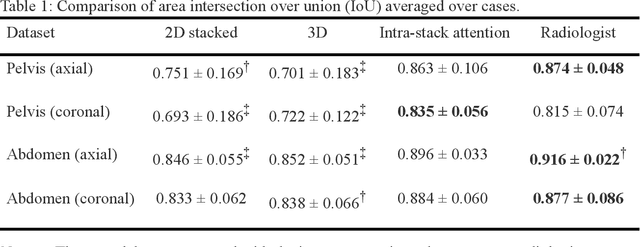
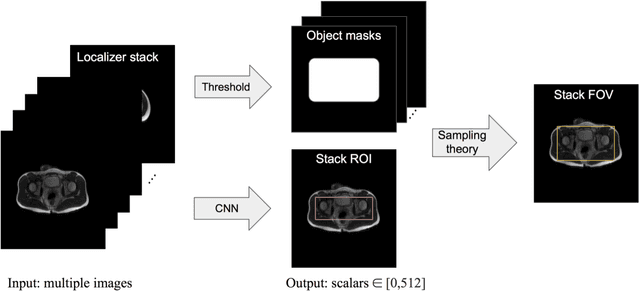
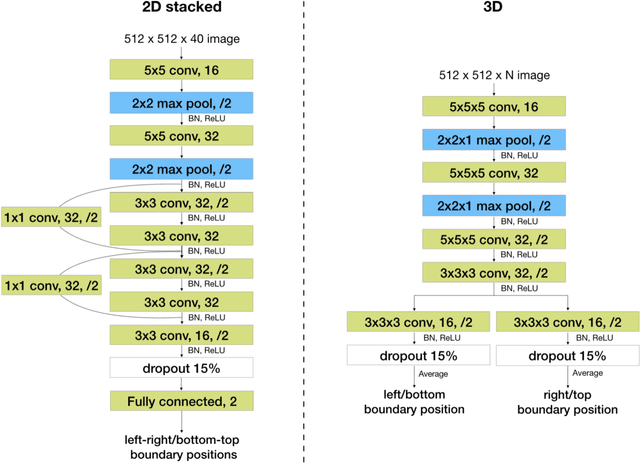
Abstract:Manual prescription of the field of view (FOV) by MRI technologists is variable and prolongs the scanning process. Often, the FOV is too large or crops critical anatomy. We propose a deep-learning framework, trained by radiologists' supervision, for automating FOV prescription. An intra-stack shared feature extraction network and an attention network are used to process a stack of 2D image inputs to generate output scalars defining the location of a rectangular region of interest (ROI). The attention mechanism is used to make the model focus on the small number of informative slices in a stack. Then the smallest FOV that makes the neural network predicted ROI free of aliasing is calculated by an algebraic operation derived from MR sampling theory. We retrospectively collected 595 cases between February 2018 and February 2022. The framework's performance is examined quantitatively with intersection over union (IoU) and pixel error on position, and qualitatively with a reader study. We use the t-test for comparing quantitative results from all models and a radiologist. The proposed model achieves an average IoU of 0.867 and average ROI position error of 9.06 out of 512 pixels on 80 test cases, significantly better (P<0.05) than two baseline models and not significantly different from a radiologist (P>0.12). Finally, the FOV given by the proposed framework achieves an acceptance rate of 92% from an experienced radiologist.
Artifact- and content-specific quality assessment for MRI with image rulers
Nov 06, 2021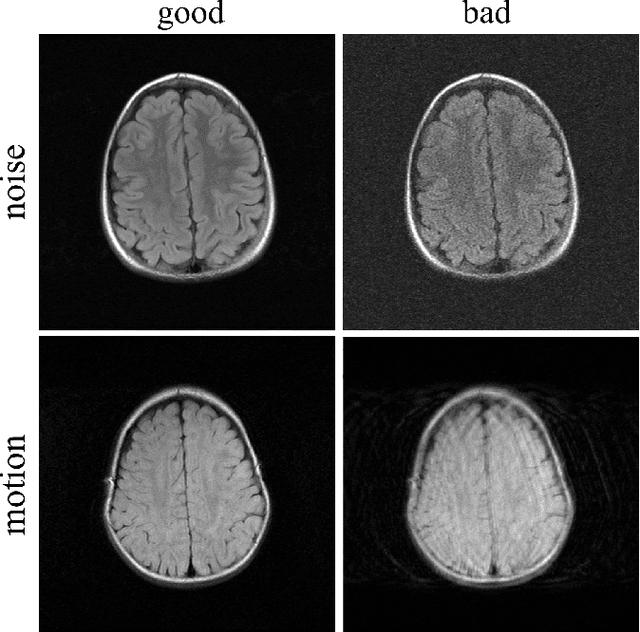
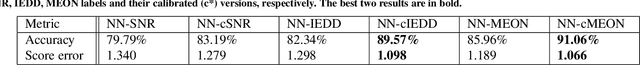
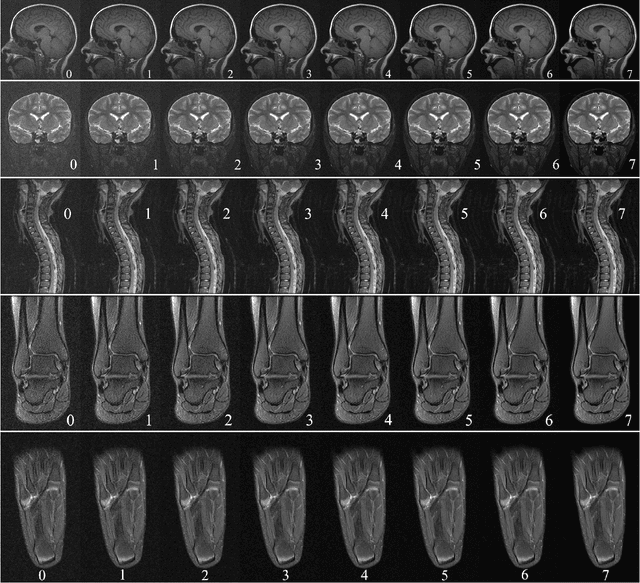
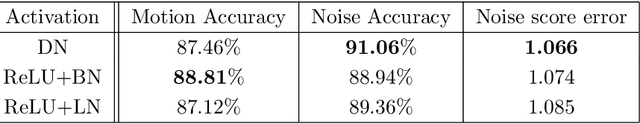
Abstract:In clinical practice MR images are often first seen by radiologists long after the scan. If image quality is inadequate either patients have to return for an additional scan, or a suboptimal interpretation is rendered. An automatic image quality assessment (IQA) would enable real-time remediation. Existing IQA works for MRI give only a general quality score, agnostic to the cause of and solution to low-quality scans. Furthermore, radiologists' image quality requirements vary with the scan type and diagnostic task. Therefore, the same score may have different implications for different scans. We propose a framework with multi-task CNN model trained with calibrated labels and inferenced with image rulers. Labels calibrated by human inputs follow a well-defined and efficient labeling task. Image rulers address varying quality standards and provide a concrete way of interpreting raw scores from the CNN. The model supports assessments of two of the most common artifacts in MRI: noise and motion. It achieves accuracies of around 90%, 6% better than the best previous method examined, and 3% better than human experts on noise assessment. Our experiments show that label calibration, image rulers, and multi-task training improve the model's performance and generalizability.
Least Squares Optimal Density Compensation for the Gridding Non-uniform Discrete Fourier Transform
Jun 16, 2021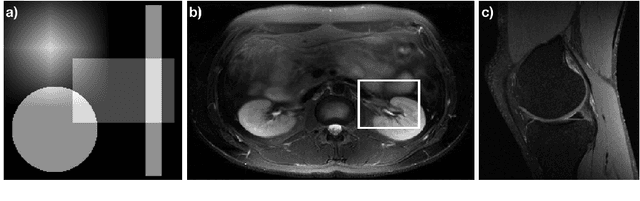
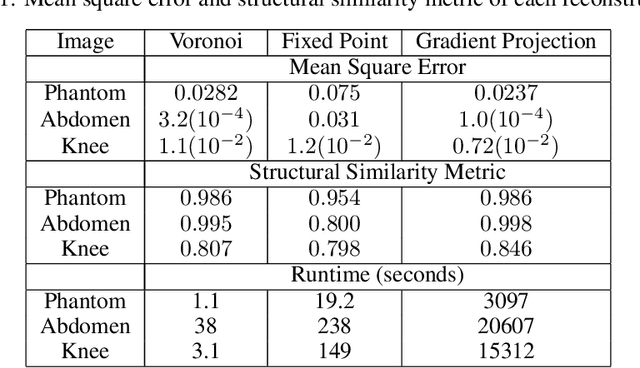
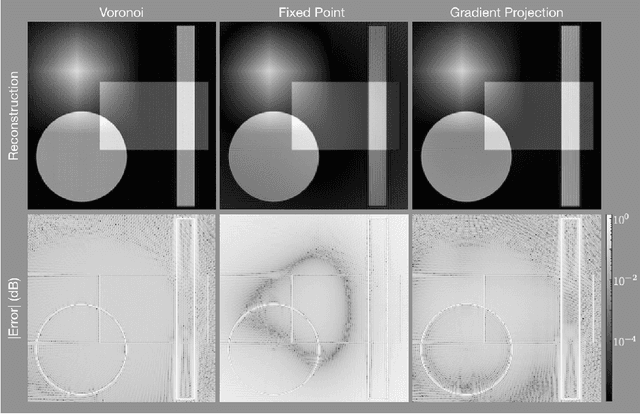
Abstract:The Gridding algorithm has shown great utility for reconstructing images from non-uniformly spaced samples in the Fourier domain in several imaging modalities. Due to the non-uniform spacing, some correction for the variable density of the samples must be made. Existing methods for generating density compensation values are either sub-optimal or only consider a finite set of points (a set of measure 0) in the optimization. This manuscript presents the first density compensation algorithm for a general trajectory that takes into account the point spread function over a set of non-zero measure. We show that the images reconstructed with Gridding using the density compensation values of this method are of superior quality when compared to density compensation weights determined in other ways. Results are shown with a numerical phantom and with magnetic resonance images of the abdomen and the knee.
Unsupervised MRI Reconstruction with Generative Adversarial Networks
Aug 29, 2020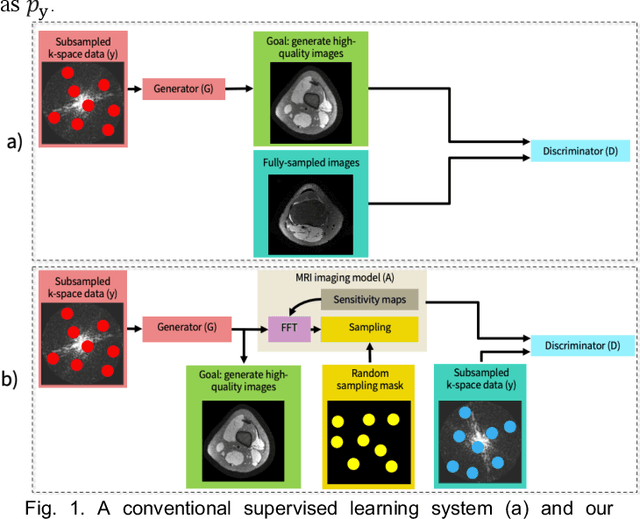
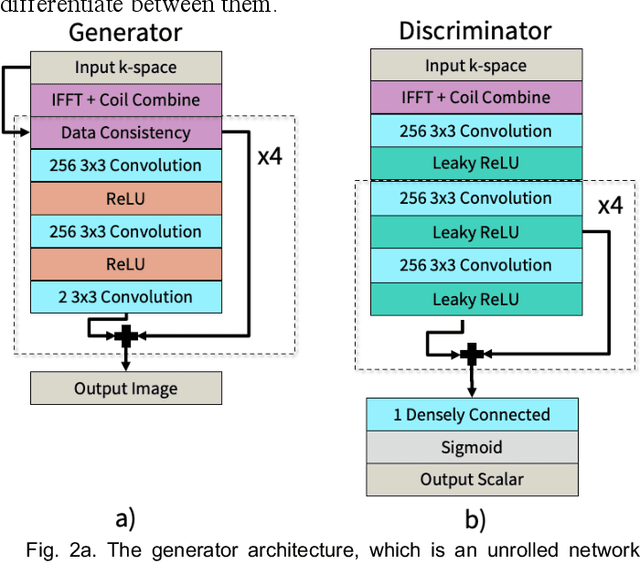
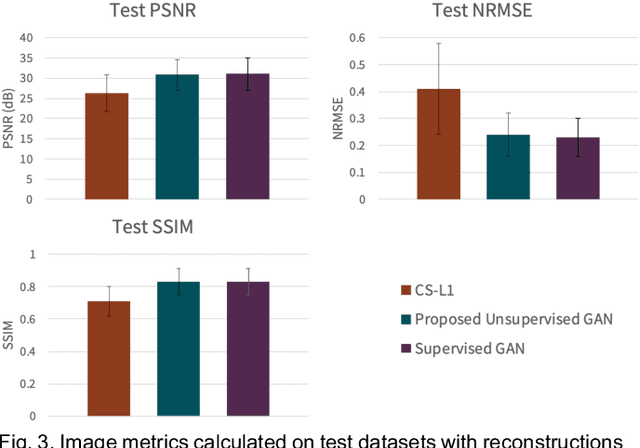
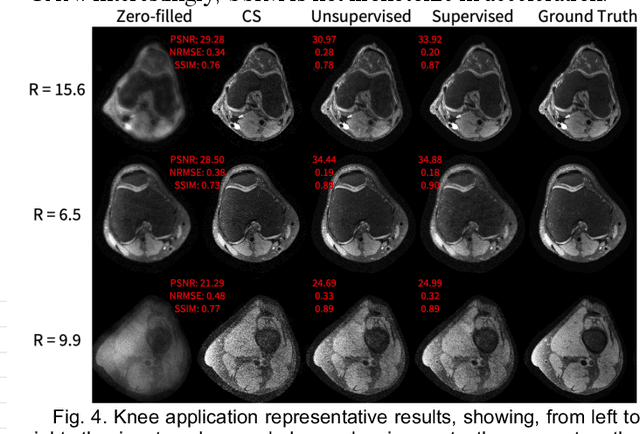
Abstract:Deep learning-based image reconstruction methods have achieved promising results across multiple MRI applications. However, most approaches require large-scale fully-sampled ground truth data for supervised training. Acquiring fully-sampled data is often either difficult or impossible, particularly for dynamic contrast enhancement (DCE), 3D cardiac cine, and 4D flow. We present a deep learning framework for MRI reconstruction without any fully-sampled data using generative adversarial networks. We test the proposed method in two scenarios: retrospectively undersampled fast spin echo knee exams and prospectively undersampled abdominal DCE. The method recovers more anatomical structure compared to conventional methods.
Multi-Domain Image Completion for Random Missing Input Data
Jul 10, 2020



Abstract:Multi-domain data are widely leveraged in vision applications taking advantage of complementary information from different modalities, e.g., brain tumor segmentation from multi-parametric magnetic resonance imaging (MRI). However, due to possible data corruption and different imaging protocols, the availability of images for each domain could vary amongst multiple data sources in practice, which makes it challenging to build a universal model with a varied set of input data. To tackle this problem, we propose a general approach to complete the random missing domain(s) data in real applications. Specifically, we develop a novel multi-domain image completion method that utilizes a generative adversarial network (GAN) with a representational disentanglement scheme to extract shared skeleton encoding and separate flesh encoding across multiple domains. We further illustrate that the learned representation in multi-domain image completion could be leveraged for high-level tasks, e.g., segmentation, by introducing a unified framework consisting of image completion and segmentation with a shared content encoder. The experiments demonstrate consistent performance improvement on three datasets for brain tumor segmentation, prostate segmentation, and facial expression image completion respectively.
Complex-Valued Convolutional Neural Networks for MRI Reconstruction
Apr 08, 2020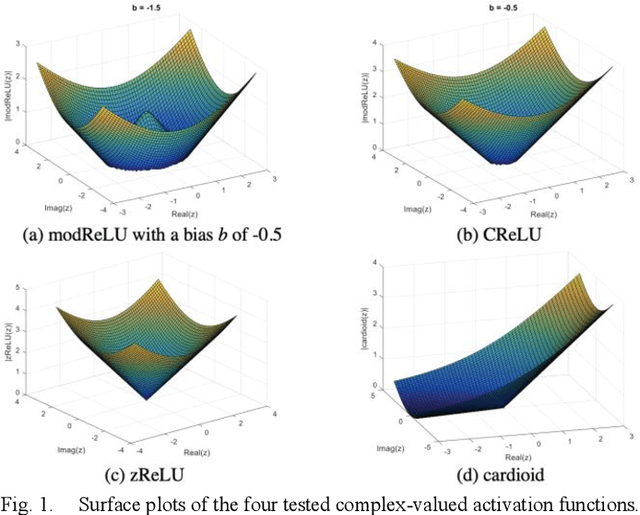
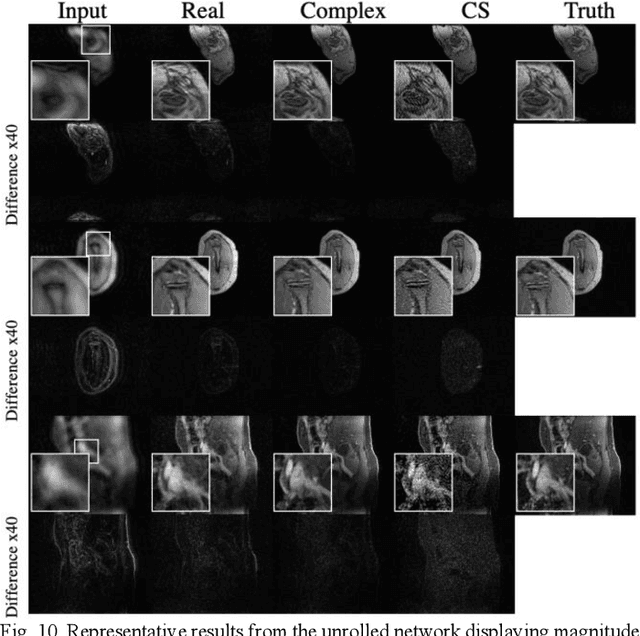
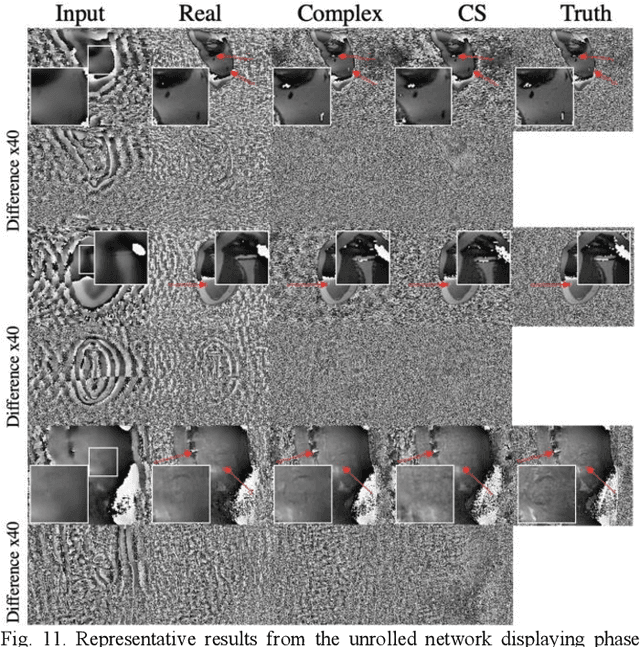
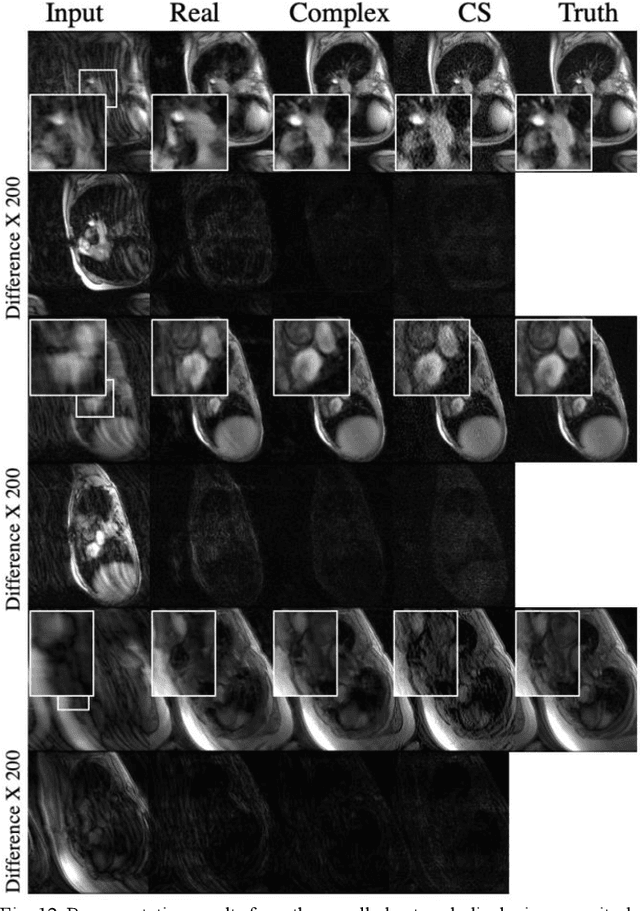
Abstract:Many real-world signal sources are complex-valued, having real and imaginary components. However, the vast majority of existing deep learning platforms and network architectures do not support the use of complex-valued data. MRI data is inherently complex-valued, so existing approaches discard the richer algebraic structure of the complex data. In this work, we investigate end-to-end complex-valued convolutional neural networks - specifically, for image reconstruction in lieu of two-channel real-valued networks. We apply this to magnetic resonance imaging reconstruction for the purpose of accelerating scan times and determine the performance of various promising complex-valued activation functions. We find that complex-valued CNNs with complex-valued convolutions provide superior reconstructions compared to real-valued convolutions with the same number of trainable parameters, over a variety of network architectures and datasets.
Diagnostic Image Quality Assessment and Classification in Medical Imaging: Opportunities and Challenges
Dec 05, 2019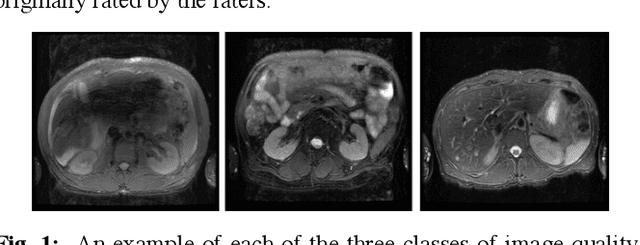
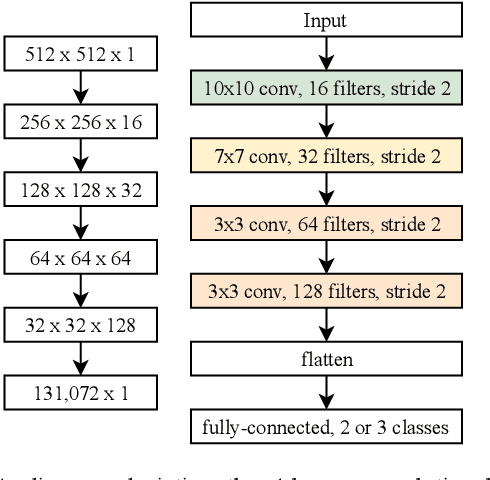
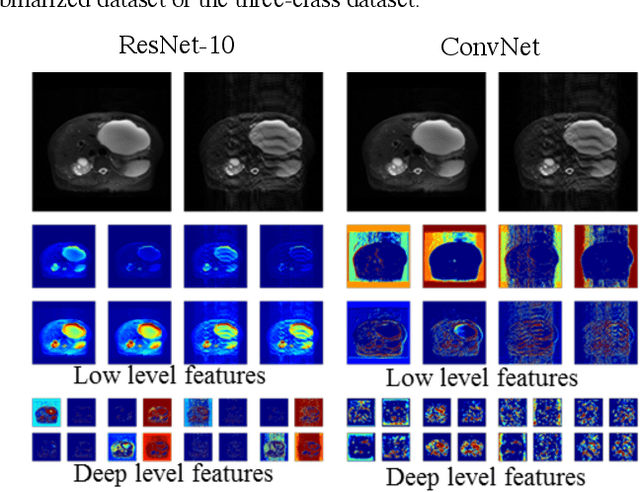
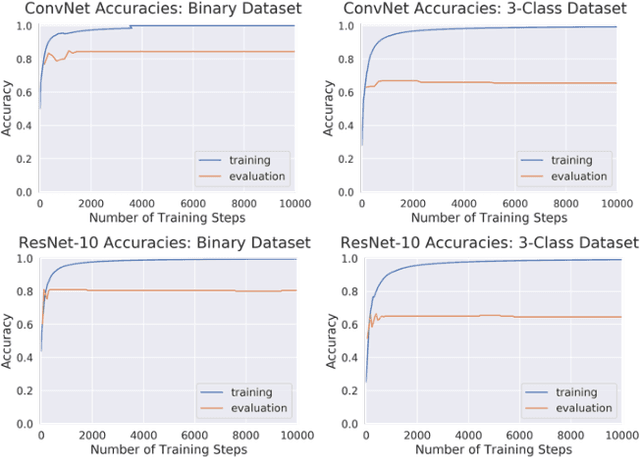
Abstract:Magnetic Resonance Imaging (MRI) suffers from several artifacts, the most common of which are motion artifacts. These artifacts often yield images that are of non-diagnostic quality. To detect such artifacts, images are prospectively evaluated by experts for their diagnostic quality, which necessitates patient-revisits and rescans whenever non-diagnostic quality scans are encountered. This motivates the need to develop an automated framework capable of accessing medical image quality and detecting diagnostic and non-diagnostic images. In this paper, we explore several convolutional neural network-based frameworks for medical image quality assessment and investigate several challenges therein.
Wasserstein GANs for MR Imaging: from Paired to Unpaired Training
Oct 15, 2019
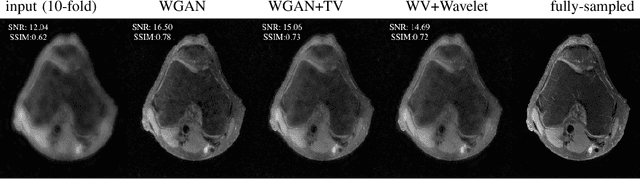
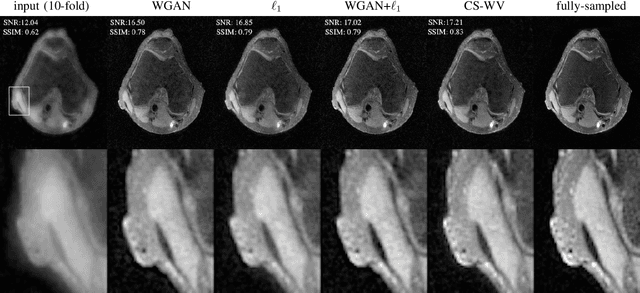
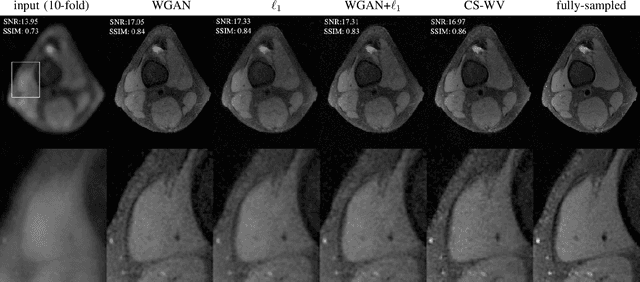
Abstract:Lack of ground-truth MR images (labels) impedes the common supervised training of deep networks for image reconstruction. To cope with this challenge, this paper leverages WGANs for unpaired training of reconstruction networks, where the inputs are the undersampled naively reconstructed images from one dataset, and the outputs are high-quality images from another dataset. The generator network is an unrolled neural network with a cascade of residual blocks and data consistency modules. The discriminator is also a multilayer CNN that plays the role of a critic scoring the quality of reconstructed images. Our extensive experiments with knee MRI datasets demonstrate unpaired WGAN training with minimal supervision is a viable option when there exists insufficient or no fully-sampled training label images that match the input images. Also, supervised paired training with additional WGAN loss achieves better and faster reconstruction compared to wavelet-based compressed sensing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge