Christopher Sandino
Learning the relative composition of EEG signals using pairwise relative shift pretraining
Nov 14, 2025Abstract:Self-supervised learning (SSL) offers a promising approach for learning electroencephalography (EEG) representations from unlabeled data, reducing the need for expensive annotations for clinical applications like sleep staging and seizure detection. While current EEG SSL methods predominantly use masked reconstruction strategies like masked autoencoders (MAE) that capture local temporal patterns, position prediction pretraining remains underexplored despite its potential to learn long-range dependencies in neural signals. We introduce PAirwise Relative Shift or PARS pretraining, a novel pretext task that predicts relative temporal shifts between randomly sampled EEG window pairs. Unlike reconstruction-based methods that focus on local pattern recovery, PARS encourages encoders to capture relative temporal composition and long-range dependencies inherent in neural signals. Through comprehensive evaluation on various EEG decoding tasks, we demonstrate that PARS-pretrained transformers consistently outperform existing pretraining strategies in label-efficient and transfer learning settings, establishing a new paradigm for self-supervised EEG representation learning.
CPEP: Contrastive Pose-EMG Pre-training Enhances Gesture Generalization on EMG Signals
Sep 04, 2025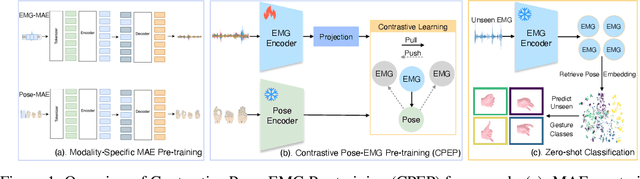
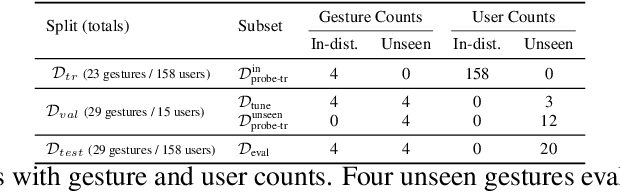
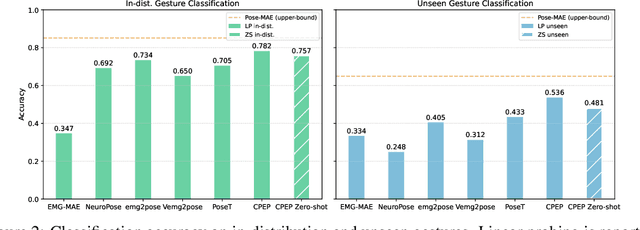
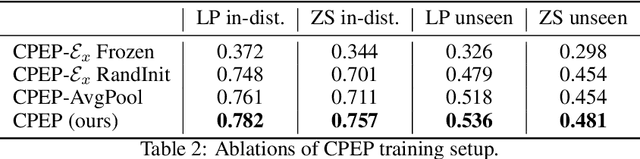
Abstract:Hand gesture classification using high-quality structured data such as videos, images, and hand skeletons is a well-explored problem in computer vision. Leveraging low-power, cost-effective biosignals, e.g. surface electromyography (sEMG), allows for continuous gesture prediction on wearables. In this paper, we demonstrate that learning representations from weak-modality data that are aligned with those from structured, high-quality data can improve representation quality and enables zero-shot classification. Specifically, we propose a Contrastive Pose-EMG Pre-training (CPEP) framework to align EMG and pose representations, where we learn an EMG encoder that produces high-quality and pose-informative representations. We assess the gesture classification performance of our model through linear probing and zero-shot setups. Our model outperforms emg2pose benchmark models by up to 21% on in-distribution gesture classification and 72% on unseen (out-of-distribution) gesture classification.
Promoting cross-modal representations to improve multimodal foundation models for physiological signals
Oct 21, 2024



Abstract:Many healthcare applications are inherently multimodal, involving several physiological signals. As sensors for these signals become more common, improving machine learning methods for multimodal healthcare data is crucial. Pretraining foundation models is a promising avenue for success. However, methods for developing foundation models in healthcare are still in early exploration and it is unclear which pretraining strategies are most effective given the diversity of physiological signals. This is partly due to challenges in multimodal health data: obtaining data across many patients is difficult and costly, there is a lot of inter-subject variability, and modalities are often heterogeneously informative across downstream tasks. Here, we explore these challenges in the PhysioNet 2018 dataset. We use a masked autoencoding objective to pretrain a multimodal model. We show that the model learns representations that can be linearly probed for a diverse set of downstream tasks. We hypothesize that cross-modal reconstruction objectives are important for successful multimodal training, as they encourage the model to integrate information across modalities. We demonstrate that modality dropout in the input space improves performance across downstream tasks. We also find that late-fusion models pretrained with contrastive learning objectives are less effective across multiple tasks. Finally, we analyze the model's representations, showing that attention weights become more cross-modal and temporally aligned with our pretraining strategy. The learned embeddings also become more distributed in terms of the modalities encoded by each unit. Overall, our work demonstrates the utility of multimodal foundation models with health data, even across diverse physiological data sources. We further argue that explicit methods for inducing cross-modality may enhance multimodal pretraining strategies.
Efficient Source-Free Time-Series Adaptation via Parameter Subspace Disentanglement
Oct 03, 2024Abstract:In this paper, we propose a framework for efficient Source-Free Domain Adaptation (SFDA) in the context of time-series, focusing on enhancing both parameter efficiency and data-sample utilization. Our approach introduces an improved paradigm for source-model preparation and target-side adaptation, aiming to enhance training efficiency during target adaptation. Specifically, we reparameterize the source model's weights in a Tucker-style decomposed manner, factorizing the model into a compact form during the source model preparation phase. During target-side adaptation, only a subset of these decomposed factors is fine-tuned, leading to significant improvements in training efficiency. We demonstrate using PAC Bayesian analysis that this selective fine-tuning strategy implicitly regularizes the adaptation process by constraining the model's learning capacity. Furthermore, this re-parameterization reduces the overall model size and enhances inference efficiency, making the approach particularly well suited for resource-constrained devices. Additionally, we demonstrate that our framework is compatible with various SFDA methods and achieves significant computational efficiency, reducing the number of fine-tuned parameters and inference overhead in terms of MACs by over 90% while maintaining model performance.
Diagnostic Image Quality Assessment and Classification in Medical Imaging: Opportunities and Challenges
Dec 05, 2019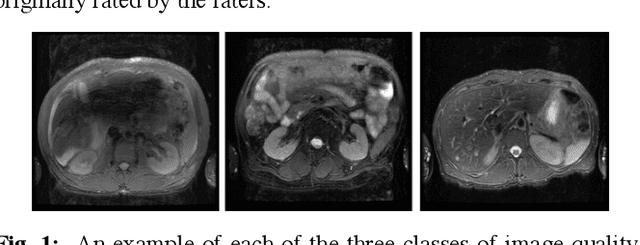
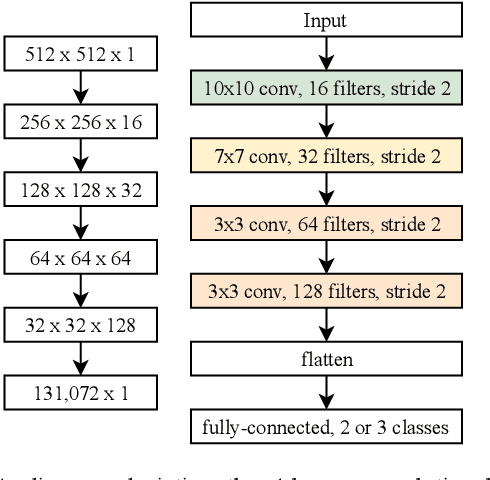
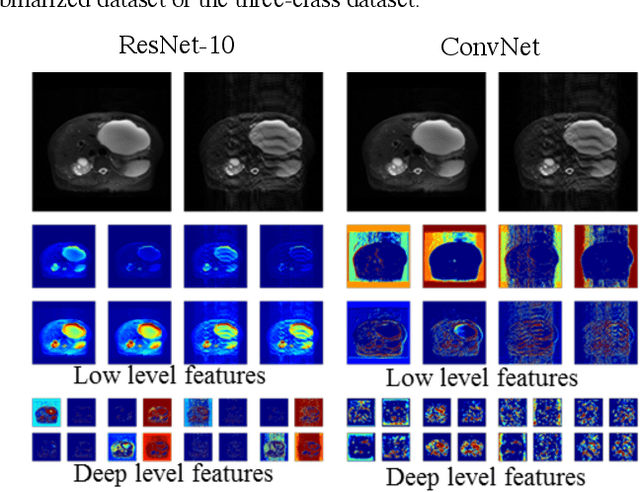
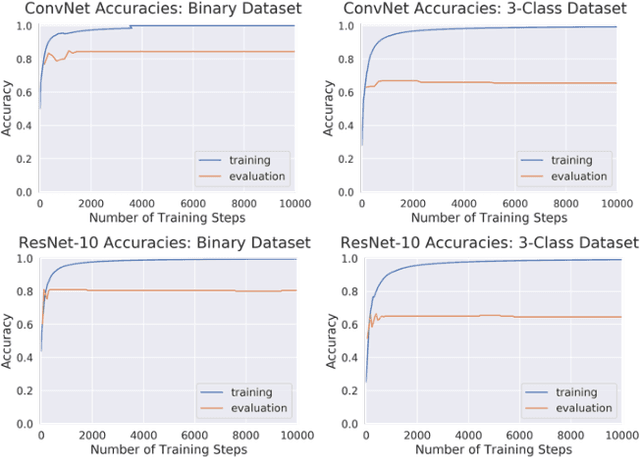
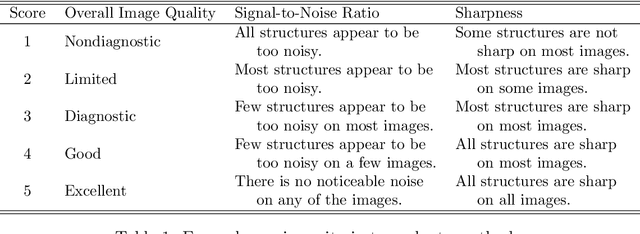
Abstract:Magnetic Resonance Imaging (MRI) suffers from several artifacts, the most common of which are motion artifacts. These artifacts often yield images that are of non-diagnostic quality. To detect such artifacts, images are prospectively evaluated by experts for their diagnostic quality, which necessitates patient-revisits and rescans whenever non-diagnostic quality scans are encountered. This motivates the need to develop an automated framework capable of accessing medical image quality and detecting diagnostic and non-diagnostic images. In this paper, we explore several convolutional neural network-based frameworks for medical image quality assessment and investigate several challenges therein.
Compressed Sensing: From Research to Clinical Practice with Data-Driven Learning
Mar 19, 2019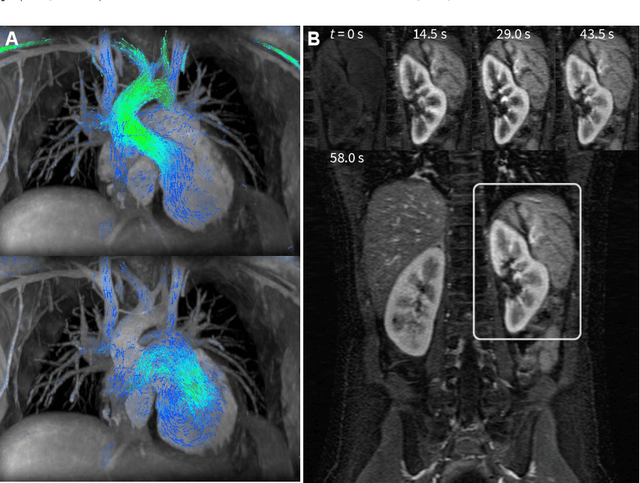
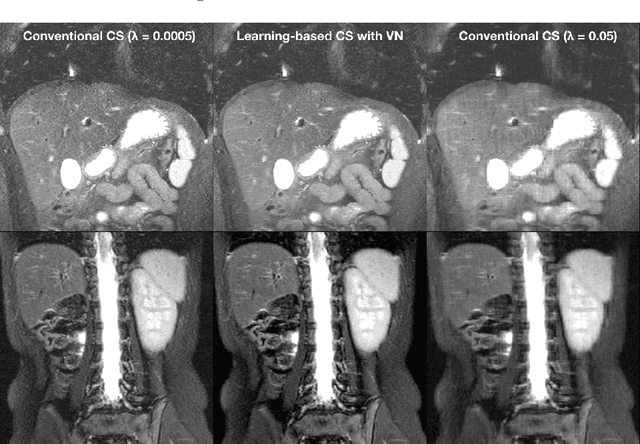
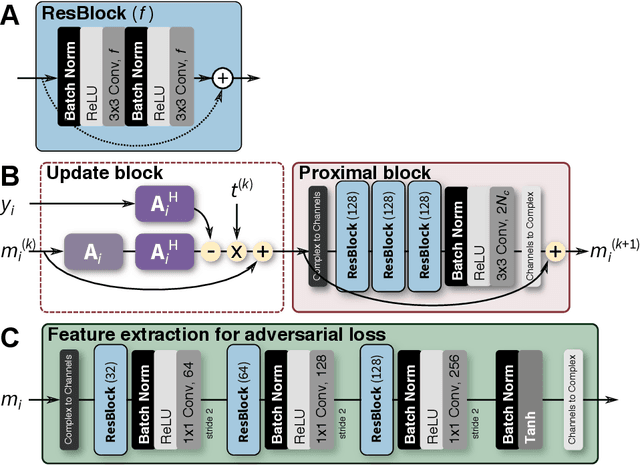
Abstract:Compressed sensing in MRI enables high subsampling factors while maintaining diagnostic image quality. This technique enables shortened scan durations and/or improved image resolution. Further, compressed sensing can increase the diagnostic information and value from each scan performed. Overall, compressed sensing has significant clinical impact in improving the diagnostic quality and patient experience for imaging exams. However, a number of challenges exist when moving compressed sensing from research to the clinic. These challenges include hand-crafted image priors, sensitive tuning parameters, and long reconstruction times. Data-driven learning provides a solution to address these challenges. As a result, compressed sensing can have greater clinical impact. In this tutorial, we will review the compressed sensing formulation and outline steps needed to transform this formulation to a deep learning framework. Supplementary open source code in python will be used to demonstrate this approach with open databases. Further, we will discuss considerations in applying data-driven compressed sensing in the clinical setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge