Ching Fang
From memories to maps: Mechanisms of in context reinforcement learning in transformers
Jun 24, 2025Abstract:Humans and animals show remarkable learning efficiency, adapting to new environments with minimal experience. This capability is not well captured by standard reinforcement learning algorithms that rely on incremental value updates. Rapid adaptation likely depends on episodic memory -- the ability to retrieve specific past experiences to guide decisions in novel contexts. Transformers provide a useful setting for studying these questions because of their ability to learn rapidly in-context and because their key-value architecture resembles episodic memory systems in the brain. We train a transformer to in-context reinforcement learn in a distribution of planning tasks inspired by rodent behavior. We then characterize the learning algorithms that emerge in the model. We first find that representation learning is supported by in-context structure learning and cross-context alignment, where representations are aligned across environments with different sensory stimuli. We next demonstrate that the reinforcement learning strategies developed by the model are not interpretable as standard model-free or model-based planning. Instead, we show that in-context reinforcement learning is supported by caching intermediate computations within the model's memory tokens, which are then accessed at decision time. Overall, we find that memory may serve as a computational resource, storing both raw experience and cached computations to support flexible behavior. Furthermore, the representations developed in the model resemble computations associated with the hippocampal-entorhinal system in the brain, suggesting that our findings may be relevant for natural cognition. Taken together, our work offers a mechanistic hypothesis for the rapid adaptation that underlies in-context learning in artificial and natural settings.
Promoting cross-modal representations to improve multimodal foundation models for physiological signals
Oct 21, 2024



Abstract:Many healthcare applications are inherently multimodal, involving several physiological signals. As sensors for these signals become more common, improving machine learning methods for multimodal healthcare data is crucial. Pretraining foundation models is a promising avenue for success. However, methods for developing foundation models in healthcare are still in early exploration and it is unclear which pretraining strategies are most effective given the diversity of physiological signals. This is partly due to challenges in multimodal health data: obtaining data across many patients is difficult and costly, there is a lot of inter-subject variability, and modalities are often heterogeneously informative across downstream tasks. Here, we explore these challenges in the PhysioNet 2018 dataset. We use a masked autoencoding objective to pretrain a multimodal model. We show that the model learns representations that can be linearly probed for a diverse set of downstream tasks. We hypothesize that cross-modal reconstruction objectives are important for successful multimodal training, as they encourage the model to integrate information across modalities. We demonstrate that modality dropout in the input space improves performance across downstream tasks. We also find that late-fusion models pretrained with contrastive learning objectives are less effective across multiple tasks. Finally, we analyze the model's representations, showing that attention weights become more cross-modal and temporally aligned with our pretraining strategy. The learned embeddings also become more distributed in terms of the modalities encoded by each unit. Overall, our work demonstrates the utility of multimodal foundation models with health data, even across diverse physiological data sources. We further argue that explicit methods for inducing cross-modality may enhance multimodal pretraining strategies.
Predictive auxiliary objectives in deep RL mimic learning in the brain
Oct 09, 2023Abstract:The ability to predict upcoming events has been hypothesized to comprise a key aspect of natural and machine cognition. This is supported by trends in deep reinforcement learning (RL), where self-supervised auxiliary objectives such as prediction are widely used to support representation learning and improve task performance. Here, we study the effects predictive auxiliary objectives have on representation learning across different modules of an RL system and how these mimic representational changes observed in the brain. We find that predictive objectives improve and stabilize learning particularly in resource-limited architectures, and we identify settings where longer predictive horizons better support representational transfer. Furthermore, we find that representational changes in this RL system bear a striking resemblance to changes in neural activity observed in the brain across various experiments. Specifically, we draw a connection between the auxiliary predictive model of the RL system and hippocampus, an area thought to learn a predictive model to support memory-guided behavior. We also connect the encoder network and the value learning network of the RL system to visual cortex and striatum in the brain, respectively. This work demonstrates how representation learning in deep RL systems can provide an interpretable framework for modeling multi-region interactions in the brain. The deep RL perspective taken here also suggests an additional role of the hippocampus in the brain -- that of an auxiliary learning system that benefits representation learning in other regions.
Biological learning in key-value memory networks
Oct 26, 2021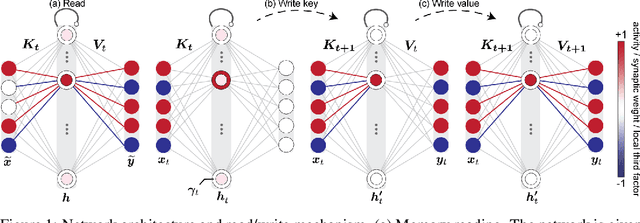


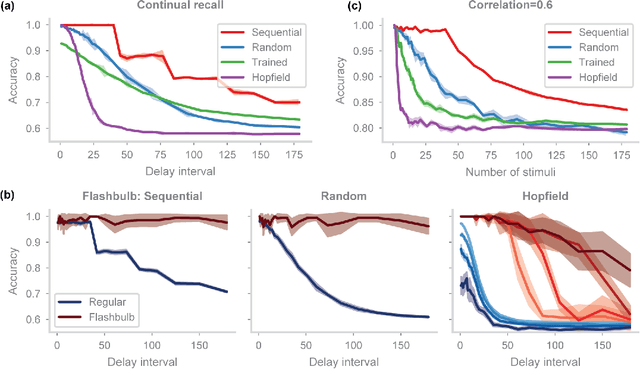
Abstract:In neuroscience, classical Hopfield networks are the standard biologically plausible model of long-term memory, relying on Hebbian plasticity for storage and attractor dynamics for recall. In contrast, memory-augmented neural networks in machine learning commonly use a key-value mechanism to store and read out memories in a single step. Such augmented networks achieve impressive feats of memory compared to traditional variants, yet their biological relevance is unclear. We propose an implementation of basic key-value memory that stores inputs using a combination of biologically plausible three-factor plasticity rules. The same rules are recovered when network parameters are meta-learned. Our network performs on par with classical Hopfield networks on autoassociative memory tasks and can be naturally extended to continual recall, heteroassociative memory, and sequence learning. Our results suggest a compelling alternative to the classical Hopfield network as a model of biological long-term memory.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge