Ali Moin
Learning the relative composition of EEG signals using pairwise relative shift pretraining
Nov 14, 2025Abstract:Self-supervised learning (SSL) offers a promising approach for learning electroencephalography (EEG) representations from unlabeled data, reducing the need for expensive annotations for clinical applications like sleep staging and seizure detection. While current EEG SSL methods predominantly use masked reconstruction strategies like masked autoencoders (MAE) that capture local temporal patterns, position prediction pretraining remains underexplored despite its potential to learn long-range dependencies in neural signals. We introduce PAirwise Relative Shift or PARS pretraining, a novel pretext task that predicts relative temporal shifts between randomly sampled EEG window pairs. Unlike reconstruction-based methods that focus on local pattern recovery, PARS encourages encoders to capture relative temporal composition and long-range dependencies inherent in neural signals. Through comprehensive evaluation on various EEG decoding tasks, we demonstrate that PARS-pretrained transformers consistently outperform existing pretraining strategies in label-efficient and transfer learning settings, establishing a new paradigm for self-supervised EEG representation learning.
Promoting cross-modal representations to improve multimodal foundation models for physiological signals
Oct 21, 2024



Abstract:Many healthcare applications are inherently multimodal, involving several physiological signals. As sensors for these signals become more common, improving machine learning methods for multimodal healthcare data is crucial. Pretraining foundation models is a promising avenue for success. However, methods for developing foundation models in healthcare are still in early exploration and it is unclear which pretraining strategies are most effective given the diversity of physiological signals. This is partly due to challenges in multimodal health data: obtaining data across many patients is difficult and costly, there is a lot of inter-subject variability, and modalities are often heterogeneously informative across downstream tasks. Here, we explore these challenges in the PhysioNet 2018 dataset. We use a masked autoencoding objective to pretrain a multimodal model. We show that the model learns representations that can be linearly probed for a diverse set of downstream tasks. We hypothesize that cross-modal reconstruction objectives are important for successful multimodal training, as they encourage the model to integrate information across modalities. We demonstrate that modality dropout in the input space improves performance across downstream tasks. We also find that late-fusion models pretrained with contrastive learning objectives are less effective across multiple tasks. Finally, we analyze the model's representations, showing that attention weights become more cross-modal and temporally aligned with our pretraining strategy. The learned embeddings also become more distributed in terms of the modalities encoded by each unit. Overall, our work demonstrates the utility of multimodal foundation models with health data, even across diverse physiological data sources. We further argue that explicit methods for inducing cross-modality may enhance multimodal pretraining strategies.
Generalizable autoregressive modeling of time series through functional narratives
Oct 10, 2024



Abstract:Time series data are inherently functions of time, yet current transformers often learn time series by modeling them as mere concatenations of time periods, overlooking their functional properties. In this work, we propose a novel objective for transformers that learn time series by re-interpreting them as temporal functions. We build an alternative sequence of time series by constructing degradation operators of different intensity in the functional space, creating augmented variants of the original sample that are abstracted or simplified to different degrees. Based on the new set of generated sequence, we train an autoregressive transformer that progressively recovers the original sample from the most simplified variant. Analogous to the next word prediction task in languages that learns narratives by connecting different words, our autoregressive transformer aims to learn the Narratives of Time Series (NoTS) by connecting different functions in time. Theoretically, we justify the construction of the alternative sequence through its advantages in approximating functions. When learning time series data with transformers, constructing sequences of temporal functions allows for a broader class of approximable functions (e.g., differentiation) compared to sequences of time periods, leading to a 26\% performance improvement in synthetic feature regression experiments. Experimentally, we validate NoTS in 3 different tasks across 22 real-world datasets, where we show that NoTS significantly outperforms other pre-training methods by up to 6\%. Additionally, combining NoTS on top of existing transformer architectures can consistently boost the performance. Our results demonstrate the potential of NoTS as a general-purpose dynamic learner, offering a viable alternative for developing foundation models for time series analysis.
Efficient Source-Free Time-Series Adaptation via Parameter Subspace Disentanglement
Oct 03, 2024Abstract:In this paper, we propose a framework for efficient Source-Free Domain Adaptation (SFDA) in the context of time-series, focusing on enhancing both parameter efficiency and data-sample utilization. Our approach introduces an improved paradigm for source-model preparation and target-side adaptation, aiming to enhance training efficiency during target adaptation. Specifically, we reparameterize the source model's weights in a Tucker-style decomposed manner, factorizing the model into a compact form during the source model preparation phase. During target-side adaptation, only a subset of these decomposed factors is fine-tuned, leading to significant improvements in training efficiency. We demonstrate using PAC Bayesian analysis that this selective fine-tuning strategy implicitly regularizes the adaptation process by constraining the model's learning capacity. Furthermore, this re-parameterization reduces the overall model size and enhances inference efficiency, making the approach particularly well suited for resource-constrained devices. Additionally, we demonstrate that our framework is compatible with various SFDA methods and achieves significant computational efficiency, reducing the number of fine-tuned parameters and inference overhead in terms of MACs by over 90% while maintaining model performance.
Frequency-Aware Masked Autoencoders for Multimodal Pretraining on Biosignals
Sep 12, 2023
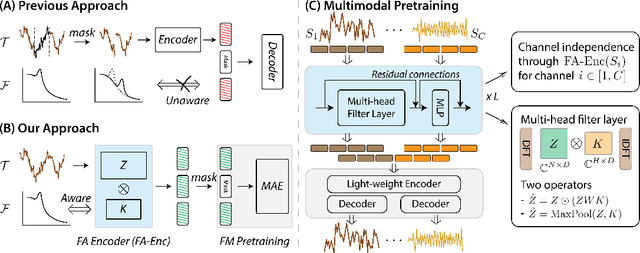
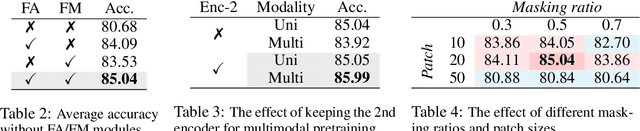
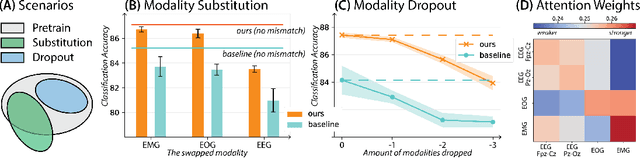
Abstract:Leveraging multimodal information from biosignals is vital for building a comprehensive representation of people's physical and mental states. However, multimodal biosignals often exhibit substantial distributional shifts between pretraining and inference datasets, stemming from changes in task specification or variations in modality compositions. To achieve effective pretraining in the presence of potential distributional shifts, we propose a frequency-aware masked autoencoder ($\texttt{bio}$FAME) that learns to parameterize the representation of biosignals in the frequency space. $\texttt{bio}$FAME incorporates a frequency-aware transformer, which leverages a fixed-size Fourier-based operator for global token mixing, independent of the length and sampling rate of inputs. To maintain the frequency components within each input channel, we further employ a frequency-maintain pretraining strategy that performs masked autoencoding in the latent space. The resulting architecture effectively utilizes multimodal information during pretraining, and can be seamlessly adapted to diverse tasks and modalities at test time, regardless of input size and order. We evaluated our approach on a diverse set of transfer experiments on unimodal time series, achieving an average of $\uparrow$5.5% improvement in classification accuracy over the previous state-of-the-art. Furthermore, we demonstrated that our architecture is robust in modality mismatch scenarios, including unpredicted modality dropout or substitution, proving its practical utility in real-world applications. Code will be available soon.
Adaptive EMG-based hand gesture recognition using hyperdimensional computing
Jan 02, 2019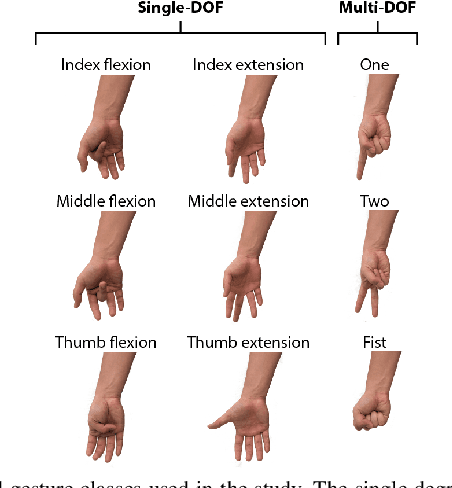


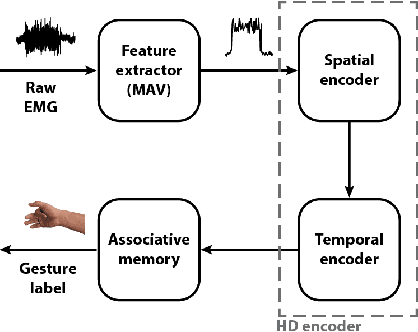
Abstract:Accurate recognition of hand gestures is crucial to the functionality of smart prosthetics and other modern human-computer interfaces. Many machine learning-based classifiers use electromyography (EMG) signals as input features, but they often misclassify gestures performed in different situational contexts (changing arm position, reapplication of electrodes, etc.) or with different effort levels due to changing signal properties. Here, we describe a learning and classification algorithm based on hyperdimensional (HD) computing that, unlike traditional machine learning algorithms, enables computationally efficient updates to incrementally incorporate new data and adapt to changing contexts. EMG signal encoding for both training and classification is performed using the same set of simple operations on 10,000-element random hypervectors enabling updates on the fly. Through human experiments using a custom EMG acquisition system, we demonstrate 88.87% classification accuracy on 13 individual finger flexion and extension gestures. Using simple model updates, we preserve this accuracy with less than 5.48% degradation when expanding to 21 commonly used gestures or when subject to changing situational contexts. We also show that the same methods for updating models can be used to account for variations resulting from the effort level with which a gesture is performed.
An EMG Gesture Recognition System with Flexible High-Density Sensors and Brain-Inspired High-Dimensional Classifier
Apr 05, 2018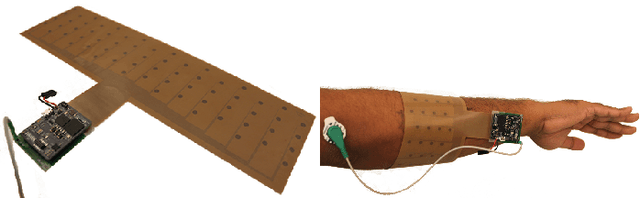
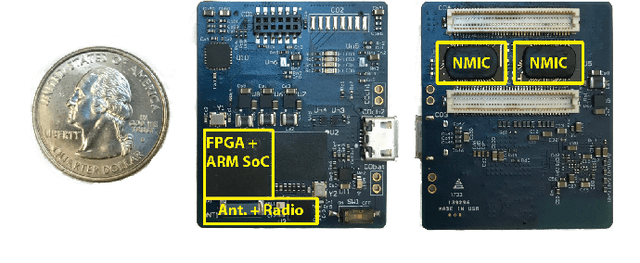
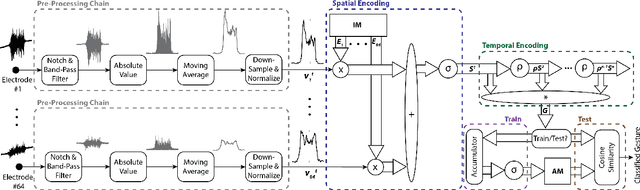
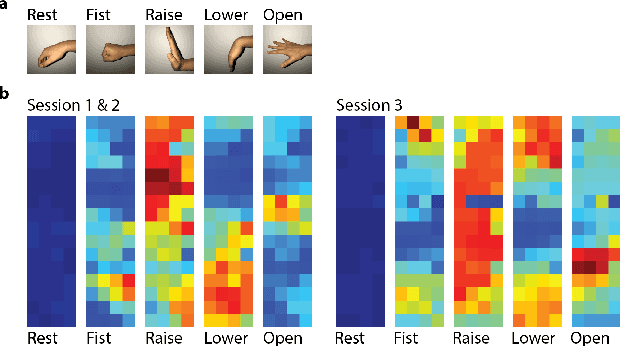
Abstract:EMG-based gesture recognition shows promise for human-machine interaction. Systems are often afflicted by signal and electrode variability which degrades performance over time. We present an end-to-end system combating this variability using a large-area, high-density sensor array and a robust classification algorithm. EMG electrodes are fabricated on a flexible substrate and interfaced to a custom wireless device for 64-channel signal acquisition and streaming. We use brain-inspired high-dimensional (HD) computing for processing EMG features in one-shot learning. The HD algorithm is tolerant to noise and electrode misplacement and can quickly learn from few gestures without gradient descent or back-propagation. We achieve an average classification accuracy of 96.64% for five gestures, with only 7% degradation when training and testing across different days. Our system maintains this accuracy when trained with only three trials of gestures; it also demonstrates comparable accuracy with the state-of-the-art when trained with one trial.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge