Frequency-Aware Masked Autoencoders for Multimodal Pretraining on Biosignals
Paper and Code
Sep 12, 2023
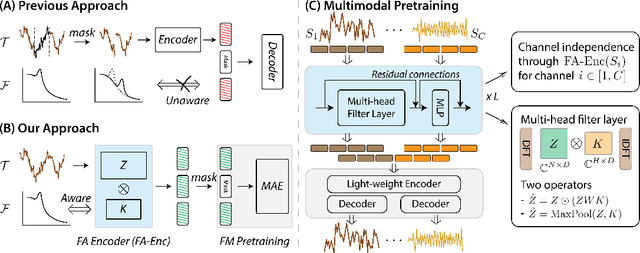
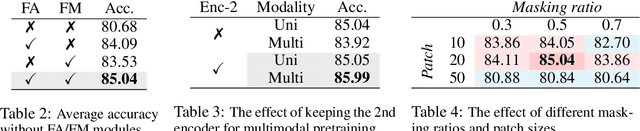
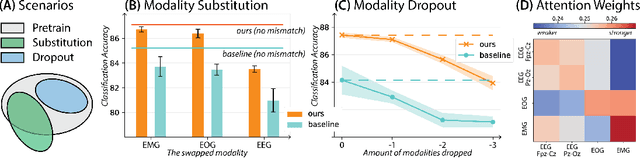
Leveraging multimodal information from biosignals is vital for building a comprehensive representation of people's physical and mental states. However, multimodal biosignals often exhibit substantial distributional shifts between pretraining and inference datasets, stemming from changes in task specification or variations in modality compositions. To achieve effective pretraining in the presence of potential distributional shifts, we propose a frequency-aware masked autoencoder ($\texttt{bio}$FAME) that learns to parameterize the representation of biosignals in the frequency space. $\texttt{bio}$FAME incorporates a frequency-aware transformer, which leverages a fixed-size Fourier-based operator for global token mixing, independent of the length and sampling rate of inputs. To maintain the frequency components within each input channel, we further employ a frequency-maintain pretraining strategy that performs masked autoencoding in the latent space. The resulting architecture effectively utilizes multimodal information during pretraining, and can be seamlessly adapted to diverse tasks and modalities at test time, regardless of input size and order. We evaluated our approach on a diverse set of transfer experiments on unimodal time series, achieving an average of $\uparrow$5.5% improvement in classification accuracy over the previous state-of-the-art. Furthermore, we demonstrated that our architecture is robust in modality mismatch scenarios, including unpredicted modality dropout or substitution, proving its practical utility in real-world applications. Code will be available soon.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge