Junshen Xu
Consistency Regularization Improves Placenta Segmentation in Fetal EPI MRI Time Series
Oct 16, 2023Abstract:The placenta plays a crucial role in fetal development. Automated 3D placenta segmentation from fetal EPI MRI holds promise for advancing prenatal care. This paper proposes an effective semi-supervised learning method for improving placenta segmentation in fetal EPI MRI time series. We employ consistency regularization loss that promotes consistency under spatial transformation of the same image and temporal consistency across nearby images in a time series. The experimental results show that the method improves the overall segmentation accuracy and provides better performance for outliers and hard samples. The evaluation also indicates that our method improves the temporal coherency of the prediction, which could lead to more accurate computation of temporal placental biomarkers. This work contributes to the study of the placenta and prenatal clinical decision-making. Code is available at https://github.com/firstmover/cr-seg.
Zero-Shot Self-Supervised Joint Temporal Image and Sensitivity Map Reconstruction via Linear Latent Space
Mar 03, 2023



Abstract:Fast spin-echo (FSE) pulse sequences for Magnetic Resonance Imaging (MRI) offer important imaging contrast in clinically feasible scan times. T2-shuffling is widely used to resolve temporal signal dynamics in FSE acquisitions by exploiting temporal correlations via linear latent space and a predefined regularizer. However, predefined regularizers fail to exploit the incoherence especially for 2D acquisitions.Recent self-supervised learning methods achieve high-fidelity reconstructions by learning a regularizer from undersampled data without a standard supervised training data set. In this work, we propose a novel approach that utilizes a self supervised learning framework to learn a regularizer constrained on a linear latent space which improves time-resolved FSE images reconstruction quality. Additionally, in regimes without groundtruth sensitivity maps, we propose joint estimation of coil-sensitivity maps using an iterative reconstruction technique. Our technique functions is in a zero-shot fashion, as it only utilizes data from a single scan of highly undersampled time series images. We perform experiments on simulated and retrospective in-vivo data to evaluate the performance of the proposed zero-shot learning method for temporal FSE reconstruction. The results demonstrate the success of our proposed method where NMSE and SSIM are significantly increased and the artifacts are reduced.
Latent Signal Models: Learning Compact Representations of Signal Evolution for Improved Time-Resolved, Multi-contrast MRI
Aug 27, 2022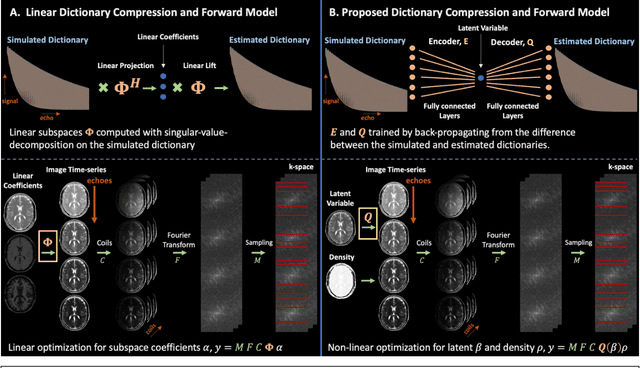
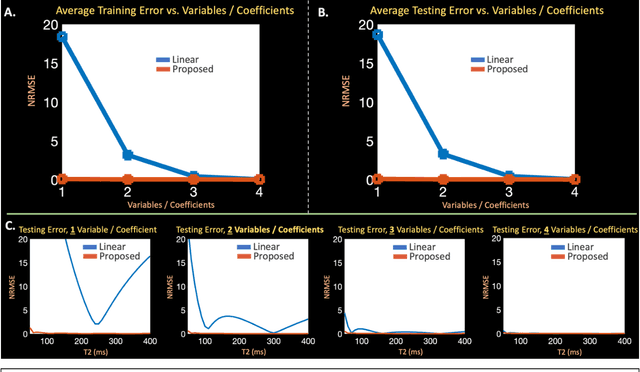
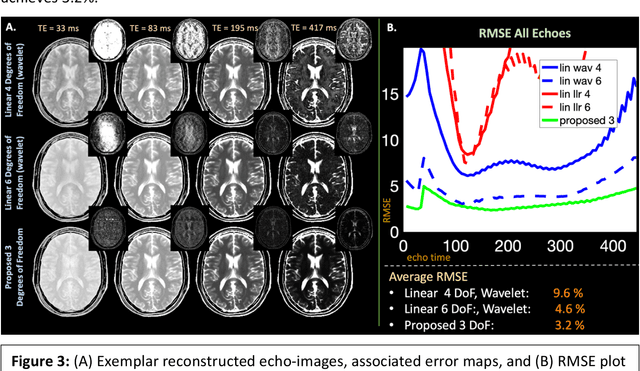
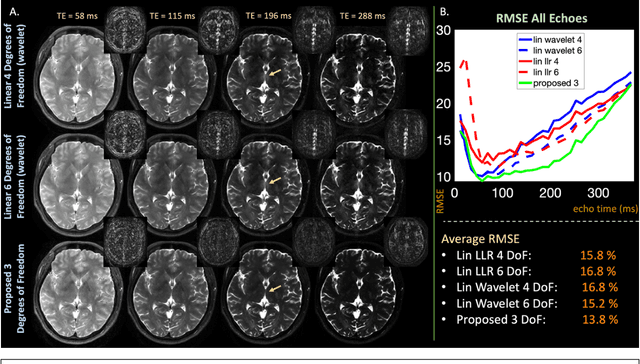
Abstract:Purpose: Training auto-encoders on simulated signal evolution and inserting the decoder into the forward model improves reconstructions through more compact, Bloch-equation-based representations of signal in comparison to linear subspaces. Methods: Building on model-based nonlinear and linear subspace techniques that enable reconstruction of signal dynamics, we train auto-encoders on dictionaries of simulated signal evolution to learn more compact, non-linear, latent representations. The proposed Latent Signal Model framework inserts the decoder portion of the auto-encoder into the forward model and directly reconstructs the latent representation. Latent Signal Models essentially serve as a proxy for fast and feasible differentiation through the Bloch-equations used to simulate signal. This work performs experiments in the context of T2-shuffling, gradient echo EPTI, and MPRAGE-shuffling. We compare how efficiently auto-encoders represent signal evolution in comparison to linear subspaces. Simulation and in-vivo experiments then evaluate if reducing degrees of freedom by inserting the decoder into the forward model improves reconstructions in comparison to subspace constraints. Results: An auto-encoder with one real latent variable represents FSE, EPTI, and MPRAGE signal evolution as well as linear subspaces characterized by four basis vectors. In simulated/in-vivo T2-shuffling and in-vivo EPTI experiments, the proposed framework achieves consistent quantitative NRMSE and qualitative improvement over linear approaches. From qualitative evaluation, the proposed approach yields images with reduced blurring and noise amplification in MPRAGE shuffling experiments. Conclusion: Directly solving for non-linear latent representations of signal evolution improves time-resolved MRI reconstructions through reduced degrees of freedom.
SVoRT: Iterative Transformer for Slice-to-Volume Registration in Fetal Brain MRI
Jun 22, 2022



Abstract:Volumetric reconstruction of fetal brains from multiple stacks of MR slices, acquired in the presence of almost unpredictable and often severe subject motion, is a challenging task that is highly sensitive to the initialization of slice-to-volume transformations. We propose a novel slice-to-volume registration method using Transformers trained on synthetically transformed data, which model multiple stacks of MR slices as a sequence. With the attention mechanism, our model automatically detects the relevance between slices and predicts the transformation of one slice using information from other slices. We also estimate the underlying 3D volume to assist slice-to-volume registration and update the volume and transformations alternately to improve accuracy. Results on synthetic data show that our method achieves lower registration error and better reconstruction quality compared with existing state-of-the-art methods. Experiments with real-world MRI data are also performed to demonstrate the ability of the proposed model to improve the quality of 3D reconstruction under severe fetal motion.
STRESS: Super-Resolution for Dynamic Fetal MRI using Self-Supervised Learning
Jun 30, 2021



Abstract:Fetal motion is unpredictable and rapid on the scale of conventional MR scan times. Therefore, dynamic fetal MRI, which aims at capturing fetal motion and dynamics of fetal function, is limited to fast imaging techniques with compromises in image quality and resolution. Super-resolution for dynamic fetal MRI is still a challenge, especially when multi-oriented stacks of image slices for oversampling are not available and high temporal resolution for recording the dynamics of the fetus or placenta is desired. Further, fetal motion makes it difficult to acquire high-resolution images for supervised learning methods. To address this problem, in this work, we propose STRESS (Spatio-Temporal Resolution Enhancement with Simulated Scans), a self-supervised super-resolution framework for dynamic fetal MRI with interleaved slice acquisitions. Our proposed method simulates an interleaved slice acquisition along the high-resolution axis on the originally acquired data to generate pairs of low- and high-resolution images. Then, it trains a super-resolution network by exploiting both spatial and temporal correlations in the MR time series, which is used to enhance the resolution of the original data. Evaluations on both simulated and in utero data show that our proposed method outperforms other self-supervised super-resolution methods and improves image quality, which is beneficial to other downstream tasks and evaluations.
Deformed2Self: Self-Supervised Denoising for Dynamic Medical Imaging
Jun 23, 2021



Abstract:Image denoising is of great importance for medical imaging system, since it can improve image quality for disease diagnosis and downstream image analyses. In a variety of applications, dynamic imaging techniques are utilized to capture the time-varying features of the subject, where multiple images are acquired for the same subject at different time points. Although signal-to-noise ratio of each time frame is usually limited by the short acquisition time, the correlation among different time frames can be exploited to improve denoising results with shared information across time frames. With the success of neural networks in computer vision, supervised deep learning methods show prominent performance in single-image denoising, which rely on large datasets with clean-vs-noisy image pairs. Recently, several self-supervised deep denoising models have been proposed, achieving promising results without needing the pairwise ground truth of clean images. In the field of multi-image denoising, however, very few works have been done on extracting correlated information from multiple slices for denoising using self-supervised deep learning methods. In this work, we propose Deformed2Self, an end-to-end self-supervised deep learning framework for dynamic imaging denoising. It combines single-image and multi-image denoising to improve image quality and use a spatial transformer network to model motion between different slices. Further, it only requires a single noisy image with a few auxiliary observations at different time frames for training and inference. Evaluations on phantom and in vivo data with different noise statistics show that our method has comparable performance to other state-of-the-art unsupervised or self-supervised denoising methods and outperforms under high noise levels.
Multi-scale Neural ODEs for 3D Medical Image Registration
Jun 17, 2021



Abstract:Image registration plays an important role in medical image analysis. Conventional optimization based methods provide an accurate estimation due to the iterative process at the cost of expensive computation. Deep learning methods such as learn-to-map are much faster but either iterative or coarse-to-fine approach is required to improve accuracy for handling large motions. In this work, we proposed to learn a registration optimizer via a multi-scale neural ODE model. The inference consists of iterative gradient updates similar to a conventional gradient descent optimizer but in a much faster way, because the neural ODE learns from the training data to adapt the gradient efficiently at each iteration. Furthermore, we proposed to learn a modal-independent similarity metric to address image appearance variations across different image contrasts. We performed evaluations through extensive experiments in the context of multi-contrast 3D MR images from both public and private data sources and demonstrate the superior performance of our proposed methods.
Enhanced detection of fetal pose in 3D MRI by Deep Reinforcement Learning with physical structure priors on anatomy
Jul 16, 2020



Abstract:Fetal MRI is heavily constrained by unpredictable and substantial fetal motion that causes image artifacts and limits the set of viable diagnostic image contrasts. Current mitigation of motion artifacts is predominantly performed by fast, single-shot MRI and retrospective motion correction. Estimation of fetal pose in real time during MRI stands to benefit prospective methods to detect and mitigate fetal motion artifacts where inferred fetal motion is combined with online slice prescription with low-latency decision making. Current developments of deep reinforcement learning (DRL), offer a novel approach for fetal landmarks detection. In this task 15 agents are deployed to detect 15 landmarks simultaneously by DRL. The optimization is challenging, and here we propose an improved DRL that incorporates priors on physical structure of the fetal body. First, we use graph communication layers to improve the communication among agents based on a graph where each node represents a fetal-body landmark. Further, additional reward based on the distance between agents and physical structures such as the fetal limbs is used to fully exploit physical structure. Evaluation of this method on a repository of 3-mm resolution in vivo data demonstrates a mean accuracy of landmark estimation within 10 mm of ground truth as 87.3%, and a mean error of 6.9 mm. The proposed DRL for fetal pose landmark search demonstrates a potential clinical utility for online detection of fetal motion that guides real-time mitigation of motion artifacts as well as health diagnosis during MRI of the pregnant mother.
Semi-Supervised Learning for Fetal Brain MRI Quality Assessment with ROI consistency
Jun 23, 2020
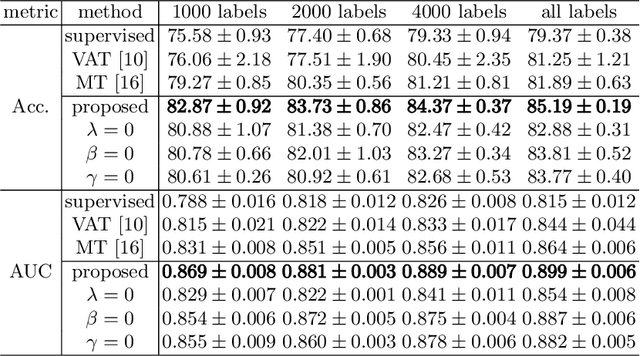
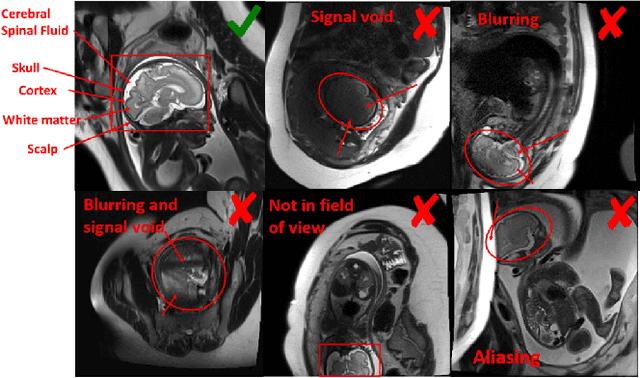
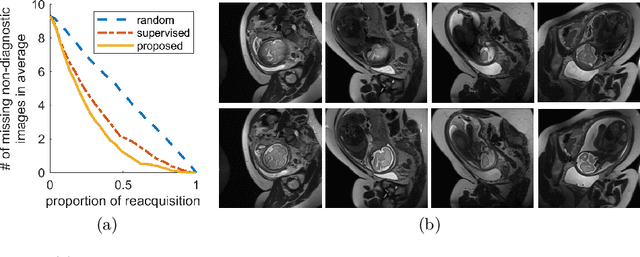
Abstract:Fetal brain MRI is useful for diagnosing brain abnormalities but is challenged by fetal motion. The current protocol for T2-weighted fetal brain MRI is not robust to motion so image volumes are degraded by inter- and intra- slice motion artifacts. Besides, manual annotation for fetal MR image quality assessment are usually time-consuming. Therefore, in this work, a semi-supervised deep learning method that detects slices with artifacts during the brain volume scan is proposed. Our method is based on the mean teacher model, where we not only enforce consistency between student and teacher models on the whole image, but also adopt an ROI consistency loss to guide the network to focus on the brain region. The proposed method is evaluated on a fetal brain MR dataset with 11,223 labeled images and more than 200,000 unlabeled images. Results show that compared with supervised learning, the proposed method can improve model accuracy by about 6\% and outperform other state-of-the-art semi-supervised learning methods. The proposed method is also implemented and evaluated on an MR scanner, which demonstrates the feasibility of online image quality assessment and image reacquisition during fetal MR scans.
Fetal Pose Estimation in Volumetric MRI using a 3D Convolution Neural Network
Jul 10, 2019



Abstract:The performance and diagnostic utility of magnetic resonance imaging (MRI) in pregnancy is fundamentally constrained by fetal motion. Motion of the fetus, which is unpredictable and rapid on the scale of conventional imaging times, limits the set of viable acquisition techniques to single-shot imaging with severe compromises in signal-to-noise ratio and diagnostic contrast, and frequently results in unacceptable image quality. Surprisingly little is known about the characteristics of fetal motion during MRI and here we propose and demonstrate methods that exploit a growing repository of MRI observations of the gravid abdomen that are acquired at low spatial resolution but relatively high temporal resolution and over long durations (10-30 minutes). We estimate fetal pose per frame in MRI volumes of the pregnant abdomen via deep learning algorithms that detect key fetal landmarks. Evaluation of the proposed method shows that our framework achieves quantitatively an average error of 4.47 mm and 96.4\% accuracy (with error less than 10 mm). Fetal pose estimation in MRI time series yields novel means of quantifying fetal movements in health and disease, and enables the learning of kinematic models that may enhance prospective mitigation of fetal motion artifacts during MRI acquisition.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge